2 Life Contingencies with R
The international section of the SOA (and the Technology section, I think!) through efforts over many years has collected the tables that are now available on the mort.soa.org. All of these tables are available in the Tables sub-directory of the shared class-notes directory in your rstudio.hpc server. So we assume your current working directory contains Tables folder, and the folder contains the files corresponding to all of the tables.
I wrote the two R function named mortalitytable and mortalitytableq that is available to you in the IntroR_Inc.R source file that you should simply source as shown below:
source("IntroR_Inc.R")These will be useful to import mortality rates from tables that are available in the XML format on http://mort.soa.org.
2.1 The List object
There are many data structures in R, of which one of them is a list. The mortalitytable returns a list object which contains not only the mortality rates but also many meta data on the table. Below, I construct and populate a list object, and then show how to access/retrieve them. If need be, use R help or the internet to understand the following piece of code.
mylist<-list(name="Shyamal",
num_children = 2,
child_ages = c(14, 14),
child_genders = c("Male","Female"),
child_nationalities = matrix(c("Mexico","US","Mexico","US"),
byrow=T, nrow=2));
mylist$name## [1] "Shyamal"mylist$num_children## [1] 2mylist$child_ages[2]## [1] 14mylist$child_nationalities[2,]## [1] "Mexico" "US"2.2 Working with an Aggregate Table
Below we will retrieve the 1980 Commissioners Standard Ordinary (CSO) Female. Basis: Age Last Birthday table, and derive some basic life contingencies quantities.
2.3 Table Retrieval and the various Data Fields of the List
mytab<-mortalitytable(35)
mytab## $number
## [1] 35
##
## $name
## [1] "1980 CSO – Female, ALB"
##
## $layout
## [1] "Aggregate"
##
## $usage
## [1] "CSO/CET"
##
## $nation
## [1] "United States of America"
##
## $select.minage
## [1] 0
##
## $select.maxage
## [1] 99
##
## $select.ageinc
## [1] 1
##
## $select.mindur
## [1] 0
##
## $select.maxdur
## [1] 100
##
## $select.durinc
## [1] 1
##
## $ult.minage
## [1] 0
##
## $ult.maxage
## [1] 99
##
## $ult.ageinc
## [1] 1
##
## $qult
## [1] 0.00188 0.00084 0.00080 0.00078 0.00077 0.00075 0.00073 0.00071 0.00070 0.00069
## [11] 0.00068 0.00070 0.00073 0.00077 0.00082 0.00087 0.00092 0.00096 0.00100 0.00103
## [21] 0.00106 0.00108 0.00110 0.00112 0.00115 0.00117 0.00120 0.00124 0.00128 0.00132
## [31] 0.00137 0.00142 0.00147 0.00154 0.00161 0.00170 0.00182 0.00196 0.00213 0.00232
## [41] 0.00253 0.00275 0.00298 0.00320 0.00344 0.00368 0.00392 0.00419 0.00448 0.00479
## [51] 0.00513 0.00550 0.00592 0.00638 0.00685 0.00733 0.00780 0.00825 0.00870 0.00920
## [61] 0.00980 0.01054 0.01149 0.01263 0.01392 0.01529 0.01671 0.01813 0.01959 0.02123
## [71] 0.02316 0.02553 0.02847 0.03199 0.03605 0.04056 0.04545 0.05068 0.05632 0.06257
## [81] 0.06967 0.07783 0.08725 0.09790 0.10962 0.12229 0.13582 0.15018 0.16538 0.18154
## [91] 0.19885 0.21768 0.23869 0.26341 0.29523 0.34102 0.41388 0.53724 0.74396 1.000002.4 Some Important Meta Data
Is it aggregate?
mytab$layout## [1] "Aggregate"When Aggregate, what is the range of the ages, and is it quinquennial or annual table?
mytab$ult.minage## [1] 0mytab$ult.maxage## [1] 99mytab$ult.ageinc # To make sure it is annual and not quinquennial## [1] 1Retrieving all the Mortality Rates
q<-mytab$qult
q## [1] 0.00188 0.00084 0.00080 0.00078 0.00077 0.00075 0.00073 0.00071 0.00070 0.00069
## [11] 0.00068 0.00070 0.00073 0.00077 0.00082 0.00087 0.00092 0.00096 0.00100 0.00103
## [21] 0.00106 0.00108 0.00110 0.00112 0.00115 0.00117 0.00120 0.00124 0.00128 0.00132
## [31] 0.00137 0.00142 0.00147 0.00154 0.00161 0.00170 0.00182 0.00196 0.00213 0.00232
## [41] 0.00253 0.00275 0.00298 0.00320 0.00344 0.00368 0.00392 0.00419 0.00448 0.00479
## [51] 0.00513 0.00550 0.00592 0.00638 0.00685 0.00733 0.00780 0.00825 0.00870 0.00920
## [61] 0.00980 0.01054 0.01149 0.01263 0.01392 0.01529 0.01671 0.01813 0.01959 0.02123
## [71] 0.02316 0.02553 0.02847 0.03199 0.03605 0.04056 0.04545 0.05068 0.05632 0.06257
## [81] 0.06967 0.07783 0.08725 0.09790 0.10962 0.12229 0.13582 0.15018 0.16538 0.18154
## [91] 0.19885 0.21768 0.23869 0.26341 0.29523 0.34102 0.41388 0.53724 0.74396 1.00000Important Check: It is important to check that the last value in the mortality rates is 1, as otherwise the resulting distribution will have mass less than \(1\).
q[length(q)]## [1] 1How to extract the rates for \((65)\)?
# Method 1 - Brute force
q<-mytab$qult;
q_65<-q[(65+1-mytab$ult.minage):length(q)];
q_65## [1] 0.01529 0.01671 0.01813 0.01959 0.02123 0.02316 0.02553 0.02847 0.03199 0.03605
## [11] 0.04056 0.04545 0.05068 0.05632 0.06257 0.06967 0.07783 0.08725 0.09790 0.10962
## [21] 0.12229 0.13582 0.15018 0.16538 0.18154 0.19885 0.21768 0.23869 0.26341 0.29523
## [31] 0.34102 0.41388 0.53724 0.74396 1.00000#Method 2 - mortalitytableq
mortalitytableq(mytab,65,0)## [1] 0.01529 0.01671 0.01813 0.01959 0.02123 0.02316 0.02553 0.02847 0.03199 0.03605
## [11] 0.04056 0.04545 0.05068 0.05632 0.06257 0.06967 0.07783 0.08725 0.09790 0.10962
## [21] 0.12229 0.13582 0.15018 0.16538 0.18154 0.19885 0.21768 0.23869 0.26341 0.29523
## [31] 0.34102 0.41388 0.53724 0.74396 1.000002.5 Computing Actuarial Quantities
First, we compute the curtate future lifetime corresponding to the above mortality table.
Method 1: Without using recurrence (Figure out why this works!)
p <- 1 - q;
kp0<-c(1,cumprod(p));
len_kp0<-length(kp0);
ex_1<-rev(cumsum(rev(kp0[-1])))/kp0[-len_kp0]Method 2 (Recurrence): Using standard recurrence
ex_2<-rep(0,mytab$ult.maxage-mytab$ult.minage+1);
for (i in (mytab$ult.maxage-mytab$ult.minage):1) {
ex_2[i]<-p[i]*(ex_2[i+1]+1);
}
ex_1[1]## [1] 74.94063ex_2[1]## [1] 74.94063Check
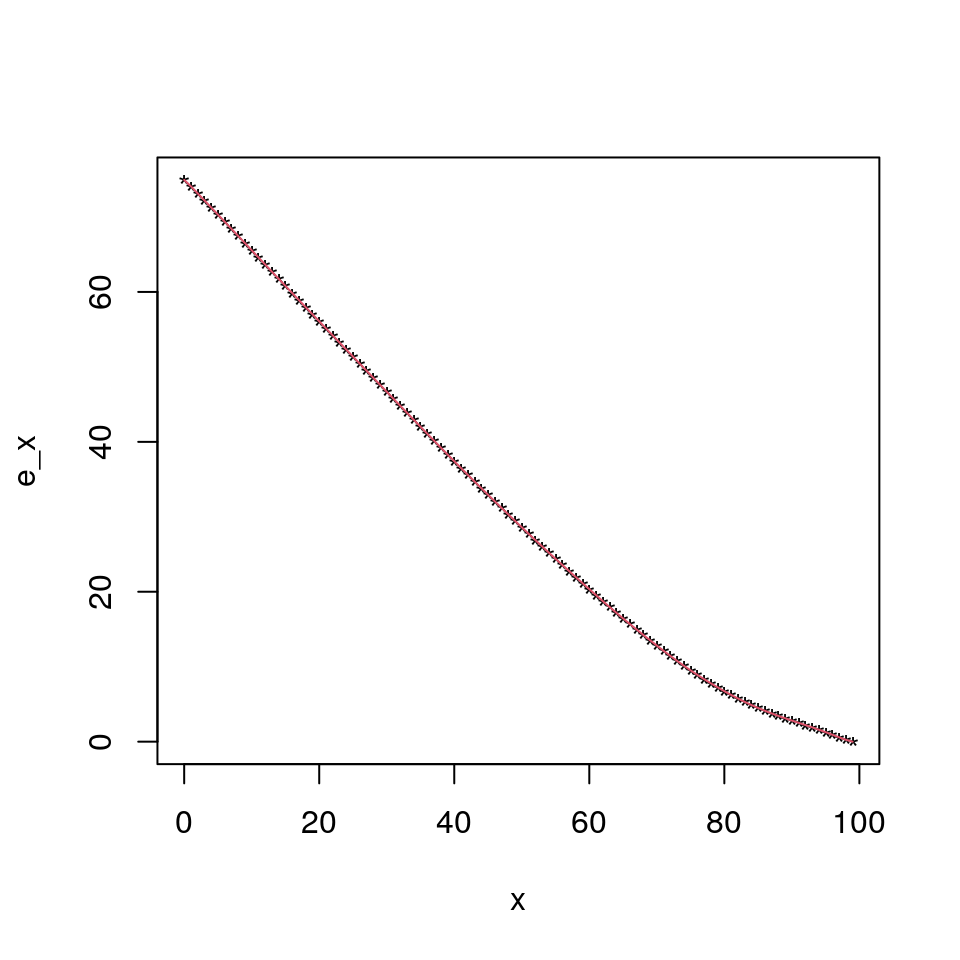
plot(0:(mytab$ult.maxage),ex_1,pch="*",ylab="e_x",xlab="x");
lines(0:(mytab$ult.maxage),ex_2,col=2);Note that for complete future lifetime under UDD you can slap a \(0.5\) to the ex’s above. Or if you wish, you could do the following:
exc<-rep(0.5,mytab$ult.maxage-mytab$ult.minage+1); # Why did I use 0.5 here?
for (i in (mytab$ult.maxage-mytab$ult.minage):1) {
exc[i]<-p[i]*(exc[i+1]+1)+(1-p[i])*0.5;
}
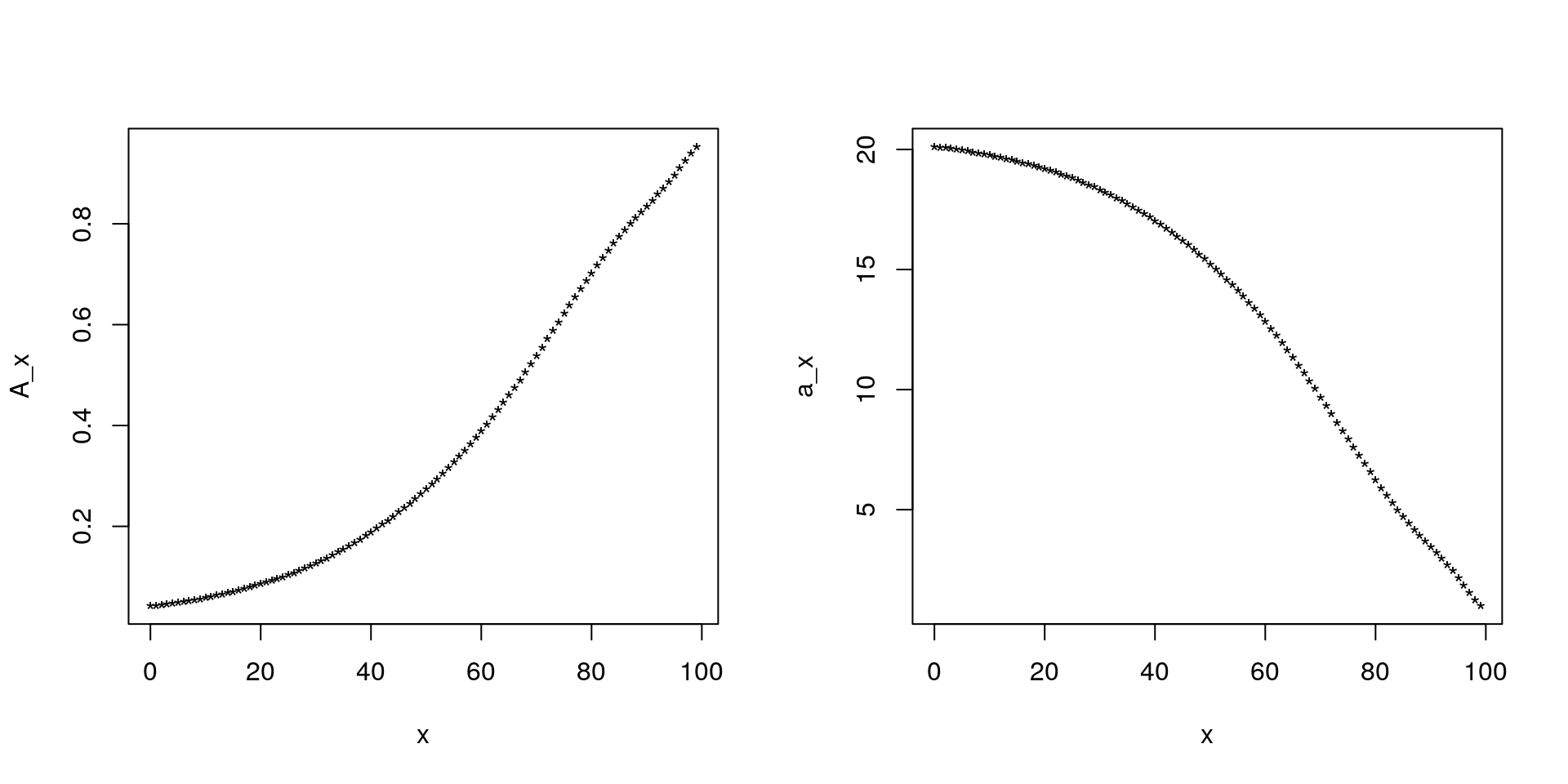
exc[1]-ex_1[1]-0.5## [1] -4.263256e-14Finally, here is how we can compute \(A_x\) and \(\ddot{a}_x\) for \(i=5\%\) :
v<-1/1.05;
Ax<-rep(v,mytab$ult.maxage-mytab$ult.minage+1); # Why did I use 0.5 here?
for (i in (mytab$ult.maxage-mytab$ult.minage):1) {
Ax[i]<-v*(p[i]*Ax[i+1]+(1-p[i]));
}
ax<-rep(1,mytab$ult.maxage-mytab$ult.minage+1); # Why did I use 1 here?
for (i in (mytab$ult.maxage-mytab$ult.minage):1) {
ax[i]<-v*p[i]*ax[i+1]+1;
}
mean((1-v)*ax+Ax) # Why should I get 1?## [1] 1par(mfrow=c(1,2));
plot(0:(mytab$ult.maxage),Ax,pch="*",ylab="A_x",xlab="x");
plot(0:(mytab$ult.maxage),ax,pch="*",ylab="a_x",xlab="x");