A Grammar of Data Manipulation
Background
The dplyr package provides a language, or grammar, for data manipulation.
The language contains a number of verbs that operate on tables in data frames.
The most commonly used verbs operate on a single data frame:
select(): pick variables by their namesfilter(): choose rows that satisfy some criteriamutate(): create transformed or derived variablesarrange(): reorder the rowssummarize(): collapse rows down to summaries
There are also a number of join verbs that merge several data frames into one.
The tidyr package provides additional verbs, such as pivot_longer() and pivot_wider() for reshaping data frames.
The single table verbs can also be used with group_by() and ungroup() to work a group at a time instead of applying to the entire data frame.
An experimental alternative to using group_by() and ungroup() is to use the .by argument with these verbs.
The design of dplyr is strongly motivated by SQL.
dplyrcan also be used to operate on tables stored in data bases.
The basic transformation verbs are described in the Data Transformation chapter of R for Data Science.
Utilities in tidyr are described in the Tidy Data chapter.
Some data sets for illustration:
EPA vehicle data used in HW4
library(readr) if (! file.exists("vehicles.csv.zip")) download.file("http://www.stat.uiowa.edu/~luke/data/vehicles.csv.zip", "vehicles.csv.zip") newmpg <- read_csv("vehicles.csv.zip", guess_max = 100000)The
nycflights13package provides data on all flights originating from one of the three main New York City airports in 2013 and heading to airports within the US.library(nycflights13)The
anyflightspackage can be used to obtain data for other cities and periods.The
stormsdata frame, included indplyrwith data on hurricanes between 1975 and 2015.
Selecting Variables
Data sets often contain hundreds of even thousands of variables.
A useful first step is to select a group of interesting variables.
A reasonable selection of the EPA vehicle variables:
newmpg1 <- select(newmpg,
make, model, year,
cty = city08, hwy = highway08,
trans = trany, cyl = cylinders,
fuel = fuelType1, displ)
head(newmpg1, 2)
## # A tibble: 2 × 9
## make model year cty hwy trans cyl fuel displ
## <chr> <chr> <dbl> <dbl> <dbl> <chr> <dbl> <chr> <dbl>
## 1 Alfa Romeo Spider Veloce 2000 1985 19 25 Manual 5-spd 4 Regu… 2
## 2 Ferrari Testarossa 1985 9 14 Manual 5-spd 12 Regu… 4.9Variables can be given new names in a select call; you can also rename them later with rename.
Some variations (the documentation for select() gives full details):
select(newmpg1, year : trans) |> head(2)
## # A tibble: 2 × 4
## year cty hwy trans
## <dbl> <dbl> <dbl> <chr>
## 1 1985 19 25 Manual 5-spd
## 2 1985 9 14 Manual 5-spdselect(newmpg1, -year, -trans) |> head(2)
## # A tibble: 2 × 7
## make model cty hwy cyl fuel displ
## <chr> <chr> <dbl> <dbl> <dbl> <chr> <dbl>
## 1 Alfa Romeo Spider Veloce 2000 19 25 4 Regular Gasoline 2
## 2 Ferrari Testarossa 9 14 12 Regular Gasoline 4.9select(newmpg1, - (year : trans)) |> head(2)
## # A tibble: 2 × 5
## make model cyl fuel displ
## <chr> <chr> <dbl> <chr> <dbl>
## 1 Alfa Romeo Spider Veloce 2000 4 Regular Gasoline 2
## 2 Ferrari Testarossa 12 Regular Gasoline 4.9Numerical column specifications can also be used:
select(newmpg1, 1, 3) |> head(2)
## # A tibble: 2 × 2
## make year
## <chr> <dbl>
## 1 Alfa Romeo 1985
## 2 Ferrari 1985select(newmpg1, 1 : 3) |> head(2)
## # A tibble: 2 × 3
## make model year
## <chr> <chr> <dbl>
## 1 Alfa Romeo Spider Veloce 2000 1985
## 2 Ferrari Testarossa 1985select(newmpg1, - (1 : 3)) |> head(2)
## # A tibble: 2 × 6
## cty hwy trans cyl fuel displ
## <dbl> <dbl> <chr> <dbl> <chr> <dbl>
## 1 19 25 Manual 5-spd 4 Regular Gasoline 2
## 2 9 14 Manual 5-spd 12 Regular Gasoline 4.9Some helper functions can also be used in the column specifications.
The most useful ones are starts_with(), ends_with(), and contains().
select(newmpg, starts_with("fuel")) |> head(2)
## # A tibble: 2 × 5
## fuelCost08 fuelCostA08 fuelType fuelType1 fuelType2
## <dbl> <dbl> <chr> <chr> <chr>
## 1 2200 0 Regular Regular Gasoline <NA>
## 2 4150 0 Regular Regular Gasoline <NA>select(newmpg, contains("city")) |> head(2)
## # A tibble: 2 × 12
## city08 city08U cityA08 cityA08U cityCD cityE cityUF rangeCity rangeCityA UCity
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 19 0 0 0 0 0 0 0 0 23.3
## 2 9 0 0 0 0 0 0 0 0 11
## # ℹ 2 more variables: UCityA <dbl>, phevCity <dbl>By default these helpers ignore case.
These can also be preceded by a minus modifier to omit columns:
select(newmpg1, -contains("m")) |> head(2)
## # A tibble: 2 × 7
## year cty hwy trans cyl fuel displ
## <dbl> <dbl> <dbl> <chr> <dbl> <chr> <dbl>
## 1 1985 19 25 Manual 5-spd 4 Regular Gasoline 2
## 2 1985 9 14 Manual 5-spd 12 Regular Gasoline 4.9contains requires a literal string; matches() is analogous but its argument is interpreted as a regular expression pattern to be matched.
select(newmpg, matches("^[Ff]uel")) |> head(2)
## # A tibble: 2 × 5
## fuelCost08 fuelCostA08 fuelType fuelType1 fuelType2
## <dbl> <dbl> <chr> <chr> <chr>
## 1 2200 0 Regular Regular Gasoline <NA>
## 2 4150 0 Regular Regular Gasoline <NA>Sometimes it is useful to move a few columns to the front; the everything() helper can be used for that.
The original NYC flights table:
head(flights, 2)
## # A tibble: 2 × 19
## year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time
## <int> <int> <int> <int> <int> <dbl> <int> <int>
## 1 2013 1 1 517 515 2 830 819
## 2 2013 1 1 533 529 4 850 830
## # ℹ 11 more variables: arr_delay <dbl>, carrier <chr>, flight <int>,
## # tailnum <chr>, origin <chr>, dest <chr>, air_time <dbl>, distance <dbl>,
## # hour <dbl>, minute <dbl>, time_hour <dttm>Moving air_time to the front:
select(flights, air_time, everything()) |> head(2)
## # A tibble: 2 × 19
## air_time year month day dep_time sched_dep_time dep_delay arr_time
## <dbl> <int> <int> <int> <int> <int> <dbl> <int>
## 1 227 2013 1 1 517 515 2 830
## 2 227 2013 1 1 533 529 4 850
## # ℹ 11 more variables: sched_arr_time <int>, arr_delay <dbl>, carrier <chr>,
## # flight <int>, tailnum <chr>, origin <chr>, dest <chr>, distance <dbl>,
## # hour <dbl>, minute <dbl>, time_hour <dttm>The where() helper function allows columns to be selected based on a predicate applied to the full column:
select(newmpg1, where(anyNA)) |> head(2)
## # A tibble: 2 × 3
## trans cyl displ
## <chr> <dbl> <dbl>
## 1 Manual 5-spd 4 2
## 2 Manual 5-spd 12 4.9select(flights, where(anyNA)) |> head(2)
## # A tibble: 2 × 6
## dep_time dep_delay arr_time arr_delay tailnum air_time
## <int> <dbl> <int> <dbl> <chr> <dbl>
## 1 517 2 830 11 N14228 227
## 2 533 4 850 20 N24211 227If there are missing values in your data it is usually a good idea to review them to make sure you understand what they mean.
Columns can also be selected using list operations:
vars <- c("make", "model", "year", "city08", "trany", "cylinders",
"fuelType1", "displ")
newmpg[vars] |> head(2)
## # A tibble: 2 × 8
## make model year city08 trany cylinders fuelType1 displ
## <chr> <chr> <dbl> <dbl> <chr> <dbl> <chr> <dbl>
## 1 Alfa Romeo Spider Veloce 2000 1985 19 Manual 5… 4 Regular … 2
## 2 Ferrari Testarossa 1985 9 Manual 5… 12 Regular … 4.9Filtering Rows
The filter() function picks out the subset of rows that satisfy one or more conditions.
All cars with city MPG at or above 140 and model year 2024:
filter(newmpg1, cty >= 140, year == 2024)
## # A tibble: 3 × 9
## make model year cty hwy trans cyl fuel displ
## <chr> <chr> <dbl> <dbl> <dbl> <chr> <dbl> <chr> <dbl>
## 1 Hyundai Ioniq 6 Long range RWD (18 … 2024 153 127 Auto… NA Elec… NA
## 2 Hyundai Ioniq 6 Standard Range RWD 2024 151 120 Auto… NA Elec… NA
## 3 Lucid Air Pure RWD with 19 inch w… 2024 140 134 Auto… NA Elec… NAAll flights from New York to Des Moines on January 1, 2013:
filter(flights, day == 1, month == 1, dest == "DSM") |>
select(year : dep_time, origin)
## # A tibble: 1 × 5
## year month day dep_time origin
## <int> <int> <int> <int> <chr>
## 1 2013 1 1 2119 EWRComparisons
The basic comparison operators are
==,!=<,<=>,>=
Be sure to use ==, not a single = character.
Be careful about floating point equality tests:
x <- 1 - 0.9
x == 0.1
## [1] FALSE
near(x, 0.1)
## [1] TRUEEquality comparisons can be used on character vectors and factors.
Order comparisons can be used on character vectors, but may produce surprising results, or results that vary with different locale settings.
%in% can be used to match against one of several options:
filter(flights, day %in% 1 : 2, month == 1, dest == "DSM") |>
select(year : dep_time, origin)
## # A tibble: 2 × 5
## year month day dep_time origin
## <int> <int> <int> <int> <chr>
## 1 2013 1 1 2119 EWR
## 2 2013 1 2 1921 EWRLogical Operations
The basic logical operations on vectors are
&– and|– or!– notxor– exclusive or
Truth tables:
x <- c(TRUE, TRUE, FALSE, FALSE)
y <- c(TRUE, FALSE, TRUE, FALSE)
! x
## [1] FALSE FALSE TRUE TRUE
x & y
## [1] TRUE FALSE FALSE FALSE
x | y
## [1] TRUE TRUE TRUE FALSE
xor(x, y)
## [1] FALSE TRUE TRUE FALSEMake sure not to confuse the vectorized logical operators & and | with the scalar flow control operators && and ||.
The previous flights example can also be written as
filter(flights, day == 1 | day == 2, month == 1, dest == "DSM") |>
select(year : dep_time, origin)
## # A tibble: 2 × 5
## year month day dep_time origin
## <int> <int> <int> <int> <chr>
## 1 2013 1 1 2119 EWR
## 2 2013 1 2 1921 EWRIt can also be written as
filter(flights, (day == 1 | day == 2) & month == 1 & dest == "DSM") |>
select(year : dep_time, origin)
## # A tibble: 2 × 5
## year month day dep_time origin
## <int> <int> <int> <int> <chr>
## 1 2013 1 1 2119 EWR
## 2 2013 1 2 1921 EWRMissing Values
filter() only keeps values corresponding to TRUE predicate values; FALSE and NA values are dropped.
Arithmetic and comparison operators will always produce NA values if one operand is NA.
1 + NA
## [1] NA
NA < 1
## [1] NA
NA == NA
## [1] NAIn a few cases a non-NA result can be determined:
TRUE | NA
## [1] TRUE
NA & FALSE
## [1] FALSE
NA ^ 0
## [1] 1is.na() can be used to also select rows with NA values.
Non-electric vehicles with zero or NA for displ:
filter(newmpg1, displ == 0 | is.na(displ), fuel != "Electricity")
## # A tibble: 2 × 9
## make model year cty hwy trans cyl fuel displ
## <chr> <chr> <dbl> <dbl> <dbl> <chr> <dbl> <chr> <dbl>
## 1 Subaru RX Turbo 1985 22 28 Manual 5-spd NA Regular Gasoline NA
## 2 Subaru RX Turbo 1985 21 27 Manual 5-spd NA Regular Gasoline NAFlights with NA for both dep_time and arr_time:
filter(flights, is.na(dep_time) & is.na(arr_time)) |>
select(year : arr_time)
## # A tibble: 8,255 × 7
## year month day dep_time sched_dep_time dep_delay arr_time
## <int> <int> <int> <int> <int> <dbl> <int>
## 1 2013 1 1 NA 1630 NA NA
## 2 2013 1 1 NA 1935 NA NA
## 3 2013 1 1 NA 1500 NA NA
## 4 2013 1 1 NA 600 NA NA
## 5 2013 1 2 NA 1540 NA NA
## 6 2013 1 2 NA 1620 NA NA
## 7 2013 1 2 NA 1355 NA NA
## 8 2013 1 2 NA 1420 NA NA
## 9 2013 1 2 NA 1321 NA NA
## 10 2013 1 2 NA 1545 NA NA
## # ℹ 8,245 more rowsAn alternative approach that does not use filter would be
idx <- with(flights, is.na(dep_time) & is.na(arr_time))
flights[idx, ] |>
select(year : arr_time)
## # A tibble: 8,255 × 7
## year month day dep_time sched_dep_time dep_delay arr_time
## <int> <int> <int> <int> <int> <dbl> <int>
## 1 2013 1 1 NA 1630 NA NA
## 2 2013 1 1 NA 1935 NA NA
## 3 2013 1 1 NA 1500 NA NA
## 4 2013 1 1 NA 600 NA NA
## 5 2013 1 2 NA 1540 NA NA
## 6 2013 1 2 NA 1620 NA NA
## 7 2013 1 2 NA 1355 NA NA
## 8 2013 1 2 NA 1420 NA NA
## 9 2013 1 2 NA 1321 NA NA
## 10 2013 1 2 NA 1545 NA NA
## # ℹ 8,245 more rowsExploring the EPA Data
Sorting out fuel types:
select(newmpg, contains("fuelType")) |> unique()
## # A tibble: 14 × 3
## fuelType fuelType1 fuelType2
## <chr> <chr> <chr>
## 1 Regular Regular Gasoline <NA>
## 2 Premium Premium Gasoline <NA>
## 3 Diesel Diesel <NA>
## 4 CNG Natural Gas <NA>
## 5 Gasoline or natural gas Regular Gasoline Natural Gas
## 6 Gasoline or E85 Regular Gasoline E85
## 7 Electricity Electricity <NA>
## 8 Gasoline or propane Regular Gasoline Propane
## 9 Premium or E85 Premium Gasoline E85
## 10 Midgrade Midgrade Gasoline <NA>
## 11 Premium Gas or Electricity Premium Gasoline Electricity
## 12 Regular Gas and Electricity Regular Gasoline Electricity
## 13 Premium and Electricity Premium Gasoline Electricity
## 14 Regular Gas or Electricity Regular Gasoline ElectricityVariables with missing values:
select(newmpg, where(anyNA)) |> head(2)
## # A tibble: 2 × 17
## cylinders displ drive eng_dscr trany guzzler trans_dscr tCharger sCharger
## <dbl> <dbl> <chr> <chr> <chr> <chr> <chr> <lgl> <chr>
## 1 4 2 Rear-Whee… (FFS) Manu… <NA> <NA> NA <NA>
## 2 12 4.9 Rear-Whee… (GUZZLE… Manu… T <NA> NA <NA>
## # ℹ 8 more variables: atvType <chr>, fuelType2 <chr>, rangeA <chr>,
## # evMotor <chr>, mfrCode <chr>, c240Dscr <chr>, c240bDscr <chr>,
## # startStop <chr>Among the reduced data set:
select(newmpg1, where(anyNA)) |> names()
## [1] "trans" "cyl" "displ"Unique missing value patterns:
incomplete_cases <- function(data) data[! complete.cases(data), ]
select(newmpg1, trans, cyl, displ, fuel) |>
incomplete_cases() |>
unique()
## # A tibble: 9 × 4
## trans cyl displ fuel
## <chr> <dbl> <dbl> <chr>
## 1 <NA> NA NA Electricity
## 2 <NA> 8 5.8 Regular Gasoline
## 3 <NA> 6 4.1 Regular Gasoline
## 4 Manual 5-spd NA NA Regular Gasoline
## 5 Manual 5-spd NA 1.3 Regular Gasoline
## 6 Automatic (A1) NA NA Electricity
## 7 Automatic (variable gear ratios) NA NA Electricity
## 8 Automatic (A1) NA 0 Electricity
## 9 Automatic (A2) NA NA ElectricityAn alternative to defining incomplete_cases():
select(newmpg1, trans, cyl, displ, fuel) |>
filter(if_any(everything(), is.na)) |>
unique()
## # A tibble: 9 × 4
## trans cyl displ fuel
## <chr> <dbl> <dbl> <chr>
## 1 <NA> NA NA Electricity
## 2 <NA> 8 5.8 Regular Gasoline
## 3 <NA> 6 4.1 Regular Gasoline
## 4 Manual 5-spd NA NA Regular Gasoline
## 5 Manual 5-spd NA 1.3 Regular Gasoline
## 6 Automatic (A1) NA NA Electricity
## 7 Automatic (variable gear ratios) NA NA Electricity
## 8 Automatic (A1) NA 0 Electricity
## 9 Automatic (A2) NA NA ElectricityLooking at the counts:
select(newmpg1, trans, cyl, displ, fuel) |>
incomplete_cases() |>
count(trans, cyl, displ, fuel)
## # A tibble: 9 × 5
## trans cyl displ fuel n
## <chr> <dbl> <dbl> <chr> <int>
## 1 Automatic (A1) NA 0 Electricity 1
## 2 Automatic (A1) NA NA Electricity 567
## 3 Automatic (A2) NA NA Electricity 63
## 4 Automatic (variable gear ratios) NA NA Electricity 9
## 5 Manual 5-spd NA 1.3 Regular Gasoline 1
## 6 Manual 5-spd NA NA Regular Gasoline 2
## 7 <NA> 6 4.1 Regular Gasoline 1
## 8 <NA> 8 5.8 Regular Gasoline 1
## 9 <NA> NA NA Electricity 9count(trans, cyl, displ, fuel) is essentially an abbreviation of
group_by(trans, cyl, displ, fuel) |>
summarize(n = n()) |>
ungroup()Another approach that avoids specifying the variables with missing values:
incomplete_cases(newmpg1) |>
count(across(where(anyNA)), fuel)
## # A tibble: 9 × 5
## trans cyl displ fuel n
## <chr> <dbl> <dbl> <chr> <int>
## 1 Automatic (A1) NA 0 Electricity 1
## 2 Automatic (A1) NA NA Electricity 567
## 3 Automatic (A2) NA NA Electricity 63
## 4 Automatic (variable gear ratios) NA NA Electricity 9
## 5 Manual 5-spd NA 1.3 Regular Gasoline 1
## 6 Manual 5-spd NA NA Regular Gasoline 2
## 7 <NA> 6 4.1 Regular Gasoline 1
## 8 <NA> 8 5.8 Regular Gasoline 1
## 9 <NA> NA NA Electricity 9Cylinders for electric vehicles:
filter(newmpg1, fuel == "Electricity") |>
count(is.na(cyl))
## # A tibble: 1 × 2
## `is.na(cyl)` n
## <lgl> <int>
## 1 TRUE 649Displacement for electric vehicles:
filter(newmpg1, fuel == "Electricity") |>
count(is.na(displ))
## # A tibble: 2 × 2
## `is.na(displ)` n
## <lgl> <int>
## 1 FALSE 1
## 2 TRUE 648Electric vehicles with non-missing displacement:
filter(newmpg1, fuel == "Electricity", ! is.na(displ))
## # A tibble: 1 × 9
## make model year cty hwy trans cyl fuel displ
## <chr> <chr> <dbl> <dbl> <dbl> <chr> <dbl> <chr> <dbl>
## 1 Mitsubishi i-MiEV 2016 126 99 Automatic (A1) NA Electricity 0Some things to think about for electric vehicles:
Should electric vehicles be dropped from analyses or visualizations involving cylinders or displacement?
Should
cylordisplbe recoded, perhaps as zero?Why are miles per gallon not missing, or infinite, for electric vehicles? The documentation for the data has some pointers.
Other considerations:
Should some transmission levels be combined?
Should some fuel type levels be combined?
Adding New Variables
mutate() can be used to define new variables or modify existing ones.
New variables are added at the end.
Later variables can be defined in terms of earlier ones.
mutate() does not modify the original data frame; it creates a new one with the specified modifications.
fl <- select(flights, year, month, day, air_time, ends_with("delay"))
mutate(fl,
gain = dep_delay - arr_delay,
air_hours = air_time / 60,
gain_per_hour = gain / air_hours) |>
head(2)
## # A tibble: 2 × 9
## year month day air_time dep_delay arr_delay gain air_hours gain_per_hour
## <int> <int> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 2013 1 1 227 2 11 -9 3.78 -2.38
## 2 2013 1 1 227 4 20 -16 3.78 -4.23transmute() keeps only the new variables.
transmute(flights,
gain = dep_delay - arr_delay,
air_hours = air_time / 60,
gain_per_hour = gain / air_hours) |>
head(2)
## # A tibble: 2 × 3
## gain air_hours gain_per_hour
## <dbl> <dbl> <dbl>
## 1 -9 3.78 -2.38
## 2 -16 3.78 -4.23Useful operators include:
Basic arithmetic operators, logarithms, etc.
Modular arithmetic operators
%/%and%%Logical operators and comparison operators.
Offset operators
lead()andlag().cumulative aggregates such as
cumsum(),cummean().Ranking operators, such as
min_rank(),percent_rank().Predicates like
is.na.
A reasonable guess is that flights with NA values for both dep_time and arr_time were cancelled:
fl <- mutate(flights,
cancelled = is.na(dep_time) & is.na(arr_time))For the geyser data set from the MASS package we used lag to obtain the duration of the previous eruption,
data(geyser, package = "MASS")
geyser2 <- mutate(geyser,
prev_duration = lag(duration))
head(geyser2, 4)
## waiting duration prev_duration
## 1 80 4.016667 NA
## 2 71 2.150000 4.016667
## 3 57 4.000000 2.150000
## 4 80 4.000000 4.000000thm <- theme_minimal() +
theme(text = element_text(size = 16))
ggplot(geyser2) +
geom_point(aes(x = prev_duration,
y = waiting)) +
thm## Warning: Removed 1 row containing missing values or values outside the scale range
## (`geom_point()`).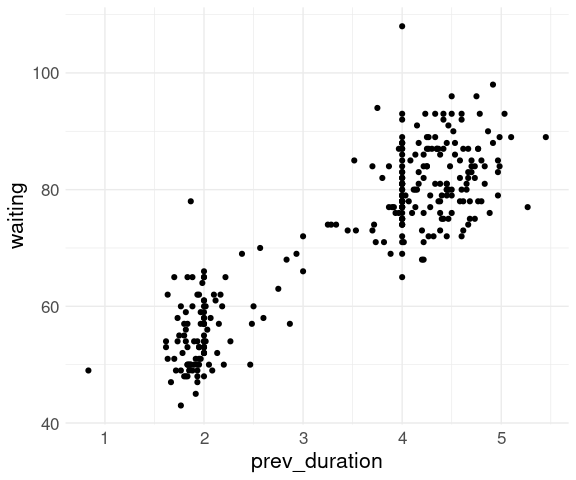
The top five vehicles in cty gas mileage (there may be more than five if there is a tie for fifth place):
mutate(newmpg1, cty_rank = min_rank(desc(cty))) |>
filter(cty_rank <= 5)
## # A tibble: 8 × 10
## make model year cty hwy trans cyl fuel displ cty_rank
## <chr> <chr> <dbl> <dbl> <dbl> <chr> <dbl> <chr> <dbl> <int>
## 1 Hyundai Ioniq Electric 2017 150 122 Auto… NA Elec… NA 5
## 2 Hyundai Ioniq Electric 2018 150 122 Auto… NA Elec… NA 5
## 3 Hyundai Ioniq Electric 2019 150 122 Auto… NA Elec… NA 5
## 4 Tesla Model 3 Standard R… 2021 150 133 Auto… NA Elec… NA 5
## 5 Hyundai Ioniq 6 Long range… 2023 153 127 Auto… NA Elec… NA 1
## 6 Hyundai Ioniq 6 Standard R… 2023 151 120 Auto… NA Elec… NA 3
## 7 Hyundai Ioniq 6 Long range… 2024 153 127 Auto… NA Elec… NA 1
## 8 Hyundai Ioniq 6 Standard R… 2024 151 120 Auto… NA Elec… NA 3An alternative to explicitly computing ranks and filtering is the slice_max() utility function:
slice_max(newmpg1, cty, n = 5)
## # A tibble: 8 × 9
## make model year cty hwy trans cyl fuel displ
## <chr> <chr> <dbl> <dbl> <dbl> <chr> <dbl> <chr> <dbl>
## 1 Hyundai Ioniq 6 Long range RWD (18 … 2023 153 127 Auto… NA Elec… NA
## 2 Hyundai Ioniq 6 Long range RWD (18 … 2024 153 127 Auto… NA Elec… NA
## 3 Hyundai Ioniq 6 Standard Range RWD 2023 151 120 Auto… NA Elec… NA
## 4 Hyundai Ioniq 6 Standard Range RWD 2024 151 120 Auto… NA Elec… NA
## 5 Hyundai Ioniq Electric 2017 150 122 Auto… NA Elec… NA
## 6 Hyundai Ioniq Electric 2018 150 122 Auto… NA Elec… NA
## 7 Hyundai Ioniq Electric 2019 150 122 Auto… NA Elec… NA
## 8 Tesla Model 3 Standard Range Plus… 2021 150 133 Auto… NA Elec… NAThe percent_rank function produces a ‘percentile rank’ (but between 0 and 1).
The top 5% of countries in life expectancy in 2007 from the gapminder data set:
library(gapminder)
gm <- filter(gapminder, year == 2007)
gm_top <- filter(gm, percent_rank(desc(lifeExp)) <= 0.05)
gm_top
## # A tibble: 8 × 6
## country continent year lifeExp pop gdpPercap
## <fct> <fct> <int> <dbl> <int> <dbl>
## 1 Australia Oceania 2007 81.2 20434176 34435.
## 2 Hong Kong, China Asia 2007 82.2 6980412 39725.
## 3 Iceland Europe 2007 81.8 301931 36181.
## 4 Israel Asia 2007 80.7 6426679 25523.
## 5 Japan Asia 2007 82.6 127467972 31656.
## 6 Spain Europe 2007 80.9 40448191 28821.
## 7 Sweden Europe 2007 80.9 9031088 33860.
## 8 Switzerland Europe 2007 81.7 7554661 37506.And the bottom 5%:
filter(gm, percent_rank(lifeExp) <= 0.05)
## # A tibble: 8 × 6
## country continent year lifeExp pop gdpPercap
## <fct> <fct> <int> <dbl> <int> <dbl>
## 1 Afghanistan Asia 2007 43.8 31889923 975.
## 2 Angola Africa 2007 42.7 12420476 4797.
## 3 Lesotho Africa 2007 42.6 2012649 1569.
## 4 Mozambique Africa 2007 42.1 19951656 824.
## 5 Sierra Leone Africa 2007 42.6 6144562 863.
## 6 Swaziland Africa 2007 39.6 1133066 4513.
## 7 Zambia Africa 2007 42.4 11746035 1271.
## 8 Zimbabwe Africa 2007 43.5 12311143 470.Within these results the original row order is retained.
The arrange function can be used to change this.
Arranging Rows
arrange() reorders the rows according to one or more variables.
By default the order is smallest to largest; using desc() reverses this.
Additional variables will be used to break ties.
Arranging the top 5% of countries in life expectancy in 2007 by life expectancy:
arrange(gm_top, desc(lifeExp))
## # A tibble: 8 × 6
## country continent year lifeExp pop gdpPercap
## <fct> <fct> <int> <dbl> <int> <dbl>
## 1 Japan Asia 2007 82.6 127467972 31656.
## 2 Hong Kong, China Asia 2007 82.2 6980412 39725.
## 3 Iceland Europe 2007 81.8 301931 36181.
## 4 Switzerland Europe 2007 81.7 7554661 37506.
## 5 Australia Oceania 2007 81.2 20434176 34435.
## 6 Spain Europe 2007 80.9 40448191 28821.
## 7 Sweden Europe 2007 80.9 9031088 33860.
## 8 Israel Asia 2007 80.7 6426679 25523.To order by continent name and order by life expectancy within continent:
arrange(gm_top, continent, desc(lifeExp))
## # A tibble: 8 × 6
## country continent year lifeExp pop gdpPercap
## <fct> <fct> <int> <dbl> <int> <dbl>
## 1 Japan Asia 2007 82.6 127467972 31656.
## 2 Hong Kong, China Asia 2007 82.2 6980412 39725.
## 3 Israel Asia 2007 80.7 6426679 25523.
## 4 Iceland Europe 2007 81.8 301931 36181.
## 5 Switzerland Europe 2007 81.7 7554661 37506.
## 6 Spain Europe 2007 80.9 40448191 28821.
## 7 Sweden Europe 2007 80.9 9031088 33860.
## 8 Australia Oceania 2007 81.2 20434176 34435.Missing values are sorted to the end:
df <- data.frame(x = c(5, 2, NA, 3))
arrange(df, x)
## x
## 1 2
## 2 3
## 3 5
## 4 NA
arrange(df, desc(x))
## x
## 1 5
## 2 3
## 3 2
## 4 NAUsing is.na you can arrange to put them at the beginning:
arrange(df, desc(is.na(x)), x)
## x
## 1 NA
## 2 2
## 3 3
## 4 5Summarizing and Grouping
summarize() collapses a table down to one row of summaries.
The average and maximal departure and arrival delays for flights, along with the total number of flights:
summarize(flights,
ave_dep_delay = mean(dep_delay, na.rm = TRUE),
ave_arr_delay = mean(arr_delay, na.rm = TRUE),
max_dep_delay = max(dep_delay, na.rm = TRUE),
max_arr_delay = max(arr_delay, na.rm = TRUE),
n = n())
## # A tibble: 1 × 5
## ave_dep_delay ave_arr_delay max_dep_delay max_arr_delay n
## <dbl> <dbl> <dbl> <dbl> <int>
## 1 12.6 6.90 1301 1272 336776Grouped Summaries
summarize() is most commonly used with grouped data to provide group level summaries.
Grouping delay summaries by destination:
fl_dest <-
group_by(flights, dest) |>
summarize(ave_dep_delay = mean(dep_delay, na.rm = TRUE),
ave_arr_delay = mean(arr_delay, na.rm = TRUE),
max_dep_delay = max(dep_delay, na.rm = TRUE),
max_arr_delay = max(arr_delay, na.rm = TRUE),
n = n()) |>
ungroup()
head(fl_dest, 5)
## # A tibble: 5 × 6
## dest ave_dep_delay ave_arr_delay max_dep_delay max_arr_delay n
## <chr> <dbl> <dbl> <dbl> <dbl> <int>
## 1 ABQ 13.7 4.38 142 153 254
## 2 ACK 6.46 4.85 219 221 265
## 3 ALB 23.6 14.4 323 328 439
## 4 ANC 12.9 -2.5 75 39 8
## 5 ATL 12.5 11.3 898 895 17215Calling ungroup is not always necessary but sometimes it is, so it is a good habit to get into.
An alternative to using group_by() and ungroup() is to use the .by argument to summarize():
flights |>
summarize(ave_dep_delay = mean(dep_delay, na.rm = TRUE),
ave_arr_delay = mean(arr_delay, na.rm = TRUE),
max_dep_delay = max(dep_delay, na.rm = TRUE),
max_arr_delay = max(arr_delay, na.rm = TRUE),
n = n(),
.by = dest) |>
arrange(dest) |> ## to match group_by() result
head()
## # A tibble: 6 × 6
## dest ave_dep_delay ave_arr_delay max_dep_delay max_arr_delay n
## <chr> <dbl> <dbl> <dbl> <dbl> <int>
## 1 ABQ 13.7 4.38 142 153 254
## 2 ACK 6.46 4.85 219 221 265
## 3 ALB 23.6 14.4 323 328 439
## 4 ANC 12.9 -2.5 75 39 8
## 5 ATL 12.5 11.3 898 895 17215
## 6 AUS 13.0 6.02 351 349 2439This is currently marked as Experimental in the documentation.
The group level summaries can be further transformed, for example to identify the top 10 destinations for average delays:
mutate(fl_dest, rank = min_rank(desc(ave_dep_delay))) |>
filter(rank <= 10)
## # A tibble: 10 × 7
## dest ave_dep_delay ave_arr_delay max_dep_delay max_arr_delay n rank
## <chr> <dbl> <dbl> <dbl> <dbl> <int> <int>
## 1 ALB 23.6 14.4 323 328 439 9
## 2 BHM 29.7 16.9 325 291 297 4
## 3 CAE 35.6 41.8 222 224 116 1
## 4 DSM 26.2 19.0 341 322 569 7
## 5 JAC 26.5 28.1 198 175 25 6
## 6 MSN 23.6 20.2 340 364 572 10
## 7 OKC 30.6 30.6 280 262 346 3
## 8 RIC 23.6 20.1 483 463 2454 8
## 9 TUL 34.9 33.7 251 262 315 2
## 10 TYS 28.5 24.1 291 281 631 5Missing Values
Most summaries will be NA if any of the data values being summarized are NA.
Typically, summary functions can be called with na.rm = TRUE to produce the summary for all non-missing values.
summarize(flights,
ave_dep_delay = mean(dep_delay, na.rm = TRUE),
ave_arr_delay = mean(arr_delay, na.rm = TRUE),
max_dep_delay = max(dep_delay, na.rm = TRUE),
max_arr_delay = max(arr_delay, na.rm = TRUE),
n = n())
## # A tibble: 1 × 5
## ave_dep_delay ave_arr_delay max_dep_delay max_arr_delay n
## <dbl> <dbl> <dbl> <dbl> <int>
## 1 12.6 6.90 1301 1272 336776You can also use filter() to remove rows with NA values in the variables to be summarized:
filter(flights, ! is.na(dep_delay), ! is.na(arr_delay)) |>
summarize(ave_dep_delay = mean(dep_delay),
ave_arr_delay = mean(arr_delay),
max_dep_delay = max(dep_delay),
max_arr_delay = max(arr_delay),
n = n())
## # A tibble: 1 × 5
## ave_dep_delay ave_arr_delay max_dep_delay max_arr_delay n
## <dbl> <dbl> <dbl> <dbl> <int>
## 1 12.6 6.90 1301 1272 327346The two approaches do produce different counts.
Counts
The helper function n() provides a count of the number of rows in the table being summarized.
Including counts with grouped summaries is usually a good idea.
Summaries based on small counts are likely to be much less reliable than ones based on larger counts.
This is reflected in higher variability among averages for destinations with fewer flights:
ggplot(fl_dest) +
geom_point(aes(x = n,
y = ave_arr_delay),
na.rm = TRUE) +
thm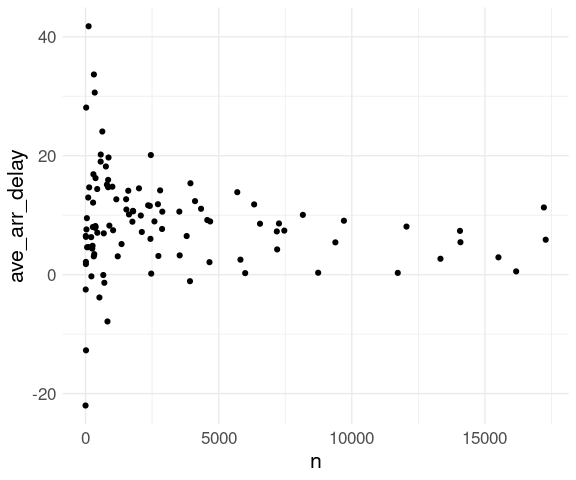
A plotly version allows the individual destinations to be identified easily:
library(plotly)
p <- ggplot(fl_dest) +
geom_point(aes(x = n,
y = ave_arr_delay,
text = dest),
na.rm = FALSE) +
thm
ggplotly(p, tooltip = "text")It would be more user-friendly to show the airport name than the three letter FAA code.
Useful Summaries
Some useful summary functions:
Location:
mean(),median()Spread:
sd(),IQR(),mad()Rank:
min(),quantile(),max()Position:
first(),nth(),last()Counts:
n().
Airline Arrival Delays
Different Delay Measures
Sometimes it is useful to look at subsets or truncated versions of a variable.
Airlines like to arrange schedules so they are often early.
ggplot(flights, aes(x = arr_delay,
y = after_stat(density))) +
geom_histogram(binwidth = 1,
na.rm = TRUE,
fill = "deepskyblue3") +
xlim(c(-100, 250)) +
thm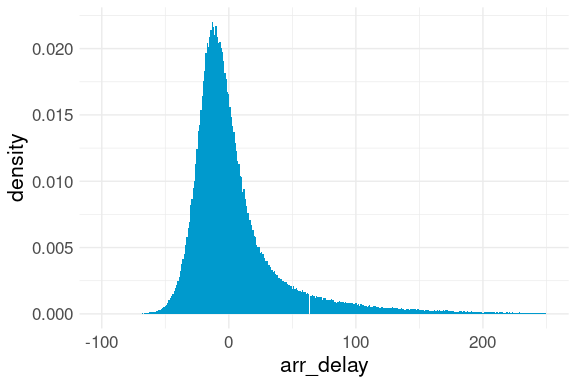
This reduces the average arrival delay but may not help travelers much.
Two options:
Consider only the positive delays for the average.
Treat negative delays as zero (zero-truncation).
The mean of the positive delays can be computed using subsetting:
mean(arr_delay[arr_delay > 0])The mean of the zero-truncated delays can be computed using the pmax function:
mean(pmax(arr_delay, 0))The summaries are
fl_arr <- filter(flights, ! is.na(arr_delay))
fl_avs <-
group_by(fl_arr, dest) |>
summarize(ave = mean(arr_delay),
ave_pos = mean(arr_delay[arr_delay > 0]),
ave_pos_zero = mean(pmax(arr_delay, 0)),
n = n()) |>
ungroup()
head(fl_avs)
## # A tibble: 6 × 5
## dest ave ave_pos ave_pos_zero n
## <chr> <dbl> <dbl> <dbl> <int>
## 1 ABQ 4.38 41.9 17.7 254
## 2 ACK 4.85 28.6 11.3 264
## 3 ALB 14.4 52.1 22.9 418
## 4 ANC -2.5 12.4 7.75 8
## 5 ATL 11.3 37.8 17.8 16837
## 6 AUS 6.02 40.6 16.6 2411One approach to visualizing the results with color coding the three different summaries:
View the
fl_avstable as a wide format withavs,ave_pos, andave_pos_zeroas three different measurement types.Use
pivot_longer()to convert to a long format with a variablewhichto hold the summary type and a variabledelayto hold the summary value:
library(tidyr)
fl <- pivot_longer(fl_avs, 2 : 4,
names_to = "which",
values_to = "delay")
arrange(fl, dest)
## # A tibble: 312 × 4
## dest n which delay
## <chr> <int> <chr> <dbl>
## 1 ABQ 254 ave 4.38
## 2 ABQ 254 ave_pos 41.9
## 3 ABQ 254 ave_pos_zero 17.7
## 4 ACK 264 ave 4.85
## 5 ACK 264 ave_pos 28.6
## 6 ACK 264 ave_pos_zero 11.3
## 7 ALB 418 ave 14.4
## 8 ALB 418 ave_pos 52.1
## 9 ALB 418 ave_pos_zero 22.9
## 10 ANC 8 ave -2.5
## # ℹ 302 more rowsA plot is then easy to create:
ggplot(fl, aes(x = n,
y = delay,
color = which)) +
geom_point() +
thm## Warning: Removed 1 row containing missing values or values outside the scale range
## (`geom_point()`).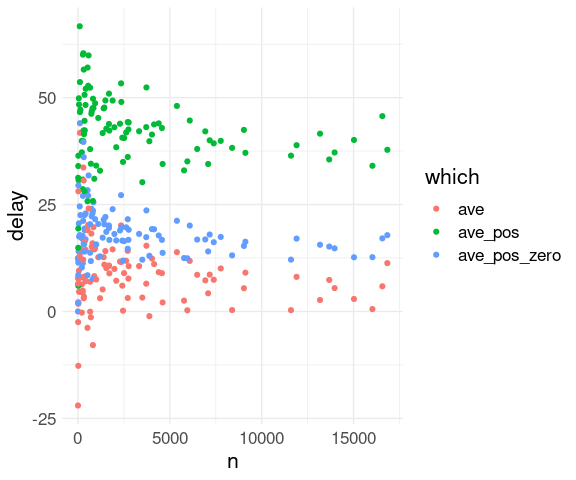
An alternative:
Use one layer per summary type.
Specify the summary type for each layer as a
coloraesthetic value.
ggplot(fl_avs, aes(x = n)) +
geom_point(aes(y = ave,
color = "ave")) +
geom_point(aes(y = ave_pos,
color = "ave_pos")) +
geom_point(aes(y = ave_pos_zero,
color = "ave_pos_zero")) +
thm## Warning: Removed 1 row containing missing values or values outside the scale range
## (`geom_point()`).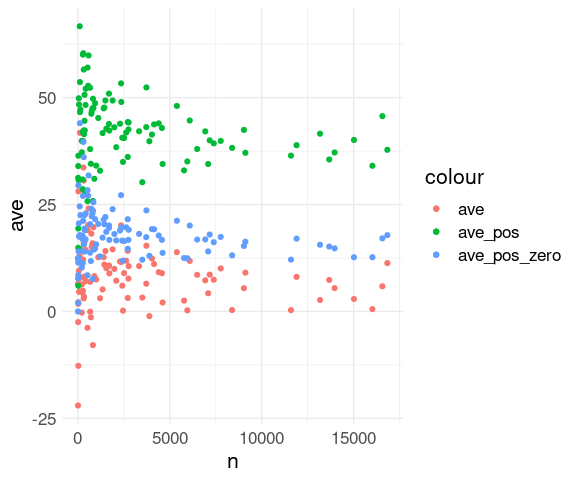
The one missing value case:
incomplete_cases(fl_avs)
## # A tibble: 1 × 5
## dest ave ave_pos ave_pos_zero n
## <chr> <dbl> <dbl> <dbl> <int>
## 1 LEX -22 NaN 0 1Delay Proportions
Proportions satisfying a condition can be computed as a mean of the logical expression for the condition:
The proportion of all flights that were delayed:
mean(fl_arr$arr_delay > 0)
## [1] 0.4063101Grouped by destination:
fl_props <-
group_by(fl_arr, dest) |>
summarize(
pd0 = mean(arr_delay > 0), ## proportion delayed
pd10 = mean(arr_delay > 10), ## more than 10 minutes
pd20 = mean(arr_delay > 20), ## more than 20 minutes
n = n()) |>
ungroup()
fl_props
## # A tibble: 104 × 5
## dest pd0 pd10 pd20 n
## <chr> <dbl> <dbl> <dbl> <int>
## 1 ABQ 0.421 0.339 0.268 254
## 2 ACK 0.394 0.246 0.167 264
## 3 ALB 0.440 0.333 0.273 418
## 4 ANC 0.625 0.125 0.125 8
## 5 ATL 0.472 0.309 0.211 16837
## 6 AUS 0.408 0.284 0.216 2411
## 7 AVL 0.456 0.257 0.207 261
## 8 BDL 0.350 0.262 0.223 412
## 9 BGR 0.383 0.299 0.246 358
## 10 BHM 0.450 0.361 0.305 269
## # ℹ 94 more rowsThese can be visualized similarly:
pivot_longer(fl_props, 2 : 4,
names_to = "which",
values_to = "prop") |>
ggplot(aes(x = n,
y = prop,
color = which)) +
geom_point() +
thm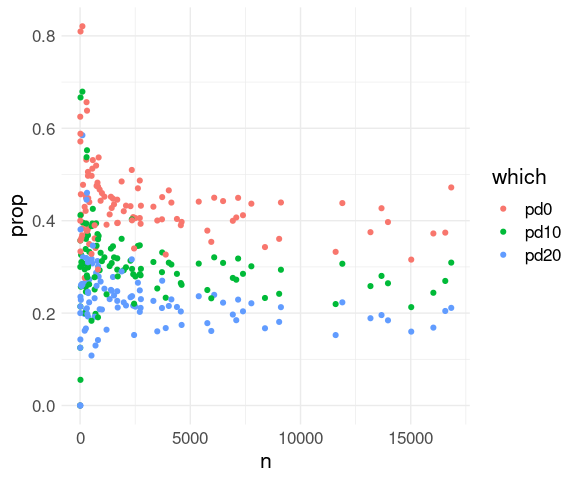
Grouped Mutate and Filter
mutate(), filter(), arrange(), slice_max(), and slice_min() can be used on a group level.
Standardizing Within Groups
mutate within groups can be used for standardizing within a group.
For example, to compute the proportion of a continent’s population living in each country for each year:
gm <- group_by(gapminder, continent, year) |>
mutate(ppop = pop / sum(pop)) |>
ungroup()Using the .by argument to mutate():
gm2 <- mutate(gapminder, ppop = pop / sum(pop), .by = c(continent, year))
identical(gm, gm2)
## [1] TRUEFor Oceania (in this data set) and years 1952 and 2007:
gm_oc <- filter(gm, continent == "Oceania", year %in% c(1952, 2007)) |>
arrange(year)
gm_oc
## # A tibble: 4 × 7
## country continent year lifeExp pop gdpPercap ppop
## <fct> <fct> <int> <dbl> <int> <dbl> <dbl>
## 1 Australia Oceania 1952 69.1 8691212 10040. 0.813
## 2 New Zealand Oceania 1952 69.4 1994794 10557. 0.187
## 3 Australia Oceania 2007 81.2 20434176 34435. 0.832
## 4 New Zealand Oceania 2007 80.2 4115771 25185. 0.168A quick check that within year and continent the proportions add up to one:
group_by(gm_oc, continent, year) |> summarize(sum_ppop = sum(ppop))
## # A tibble: 2 × 3
## # Groups: continent [1]
## continent year sum_ppop
## <fct> <int> <dbl>
## 1 Oceania 1952 1
## 2 Oceania 2007 1Focus on the 2 countries in each continent with the highest proportions of the continent’s population:
gmt2 <- group_by(gm, continent, year) |>
slice_max(ppop, n = 2) |>
ungroup() |>
filter(continent != "Oceania") ## drop as there are only two
ggplot(gmt2, aes(x = year, y = ppop,
group = country, color = country)) +
geom_line() + geom_point() +
facet_wrap(~ continent, scales = "free_y") +
thm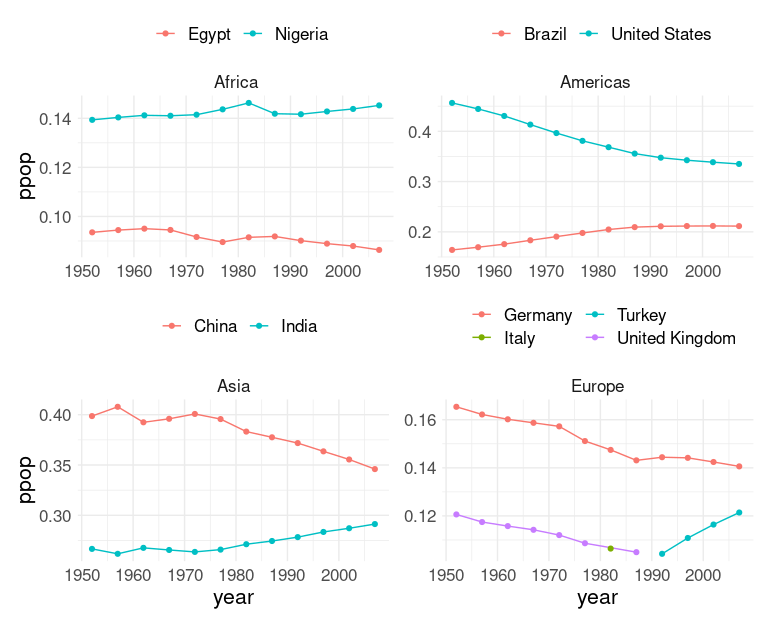
Another approach:
Create separate plots for each continent.
Adjust the themes a little.
Use
facet_wrap()to get the facet label.Arrange the plots, e.g. using
patchworkoperators.
A useful feature is that the %+% operator can be used to change the base data frame for a plot:
pAf <- ggplot(filter(gmt2, continent == "Africa"),
aes(x = year, y = ppop, col = country)) +
geom_line() + geom_point() +
facet_wrap(~ continent) +
thm +
theme(legend.position = "top",
legend.title = element_blank())
thm_nx <- theme(axis.title.x = element_blank())
thm_ny <- theme(axis.title.y = element_blank())
pAm <- pAf %+% filter(gmt2, continent == "Americas")
pAs <- pAf %+% filter(gmt2, continent == "Asia")
pEu <- pAf %+% filter(gmt2, continent == "Europe") +
guides(color = guide_legend(nrow = 2))
library(patchwork)
((pAf + thm_nx) | (pAm + thm_nx + thm_ny)) /
(pAs | (pEu + thm_ny))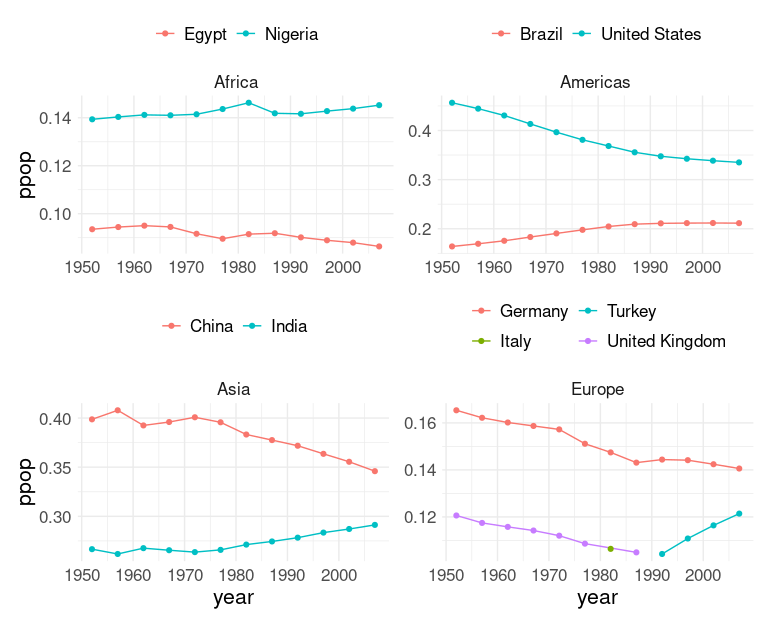
Changes Over Time
Changes over time within a country can also be examined with a grouped mutate and the lag operator.
For variables collected over time the lag function can be used to compute the previous value of a variable:
gm_us <- filter(gapminder,
country == "United States") |>
transmute(year,
lifeExp,
prev_lifeExp = lag(lifeExp))
gm_us
## # A tibble: 12 × 3
## year lifeExp prev_lifeExp
## <int> <dbl> <dbl>
## 1 1952 68.4 NA
## 2 1957 69.5 68.4
## 3 1962 70.2 69.5
## 4 1967 70.8 70.2
## 5 1972 71.3 70.8
## 6 1977 73.4 71.3
## 7 1982 74.6 73.4
## 8 1987 75.0 74.6
## 9 1992 76.1 75.0
## 10 1997 76.8 76.1
## 11 2002 77.3 76.8
## 12 2007 78.2 77.3This can then be used to compute the change from one period to the next:
mutate(gm_us,
le_change = lifeExp - prev_lifeExp)
## # A tibble: 12 × 4
## year lifeExp prev_lifeExp le_change
## <int> <dbl> <dbl> <dbl>
## 1 1952 68.4 NA NA
## 2 1957 69.5 68.4 1.05
## 3 1962 70.2 69.5 0.720
## 4 1967 70.8 70.2 0.550
## 5 1972 71.3 70.8 0.580
## 6 1977 73.4 71.3 2.04
## 7 1982 74.6 73.4 1.27
## 8 1987 75.0 74.6 0.370
## 9 1992 76.1 75.0 1.07
## 10 1997 76.8 76.1 0.720
## 11 2002 77.3 76.8 0.5
## 12 2007 78.2 77.3 0.932Use this idea to compute changes in lifeExp and gdpPercap for all countries in a grouped mutate:
gm <- group_by(gapminder, country) |>
mutate(
le_change = lifeExp - lag(lifeExp),
gdp_change = gdpPercap - lag(gdpPercap),
gdp_pct_change =
100 * gdp_change / lag(gdpPercap)) |>
ungroup()slice_max() can then be applied to data grouped by country to extract the rows with the highest gdp_pct_change for each country:
group_by(gm, country) |>
slice_max(gdp_pct_change, n = 1) |>
ungroup() |>
select(1 : 3, contains("gdp"))
## # A tibble: 142 × 6
## country continent year gdpPercap gdp_change gdp_pct_change
## <fct> <fct> <int> <dbl> <dbl> <dbl>
## 1 Afghanistan Asia 2007 975. 248. 34.1
## 2 Albania Europe 2002 4604. 1411. 44.2
## 3 Algeria Africa 1972 4183. 936. 28.8
## 4 Angola Africa 2007 4797. 2024. 73.0
## 5 Argentina Americas 2007 12779. 3982. 45.3
## 6 Australia Oceania 1967 14526. 2309. 18.9
## 7 Austria Europe 1957 8843. 2706. 44.1
## 8 Bahrain Asia 2007 29796. 6392. 27.3
## 9 Bangladesh Asia 2007 1391. 255. 22.4
## 10 Belgium Europe 1972 16672. 3523. 26.8
## # ℹ 132 more rowsGrouping by continent allows the worst drop in life expectancy for each continent to be found:
group_by(gm, continent) |>
slice_min(le_change, n = 1) |>
ungroup() |>
select(-contains("gdp"))
## # A tibble: 5 × 6
## country continent year lifeExp pop le_change
## <fct> <fct> <int> <dbl> <int> <dbl>
## 1 Rwanda Africa 1992 23.6 7290203 -20.4
## 2 El Salvador Americas 1977 56.7 4282586 -1.51
## 3 Cambodia Asia 1977 31.2 6978607 -9.10
## 4 Montenegro Europe 2002 74.0 720230 -1.46
## 5 Australia Oceania 1967 71.1 11872264 0.170Without grouping, the row with best percent GDP change overall can be computed:
slice_max(gm, gdp_pct_change, n = 1) |>
select(country, continent, year, gdp_pct_change)
## # A tibble: 1 × 4
## country continent year gdp_pct_change
## <fct> <fct> <int> <dbl>
## 1 Libya Africa 1967 178.The computation of the best GDP growth years by country can be combined into a single pipe:
group_by(gapminder, country) |>
mutate(gdp_change = gdpPercap - lag(gdpPercap),
gdp_pct_change = 100 * gdp_change / lag(gdpPercap)) |>
ungroup() |>
group_by(country) |>
slice_max(gdp_pct_change, n = 1) |>
ungroup() |>
select(1 : 3, contains("gdp"))Since the groupings of the two steps are the same and grouped mutate preserves the group structure, the middle ungroup/group_by can be dropped:
group_by(gapminder, country) |>
mutate(gdp_change = gdpPercap - lag(gdpPercap),
gdp_pct_change = 100 * gdp_change / lag(gdpPercap)) |>
slice_max(gdp_pct_change, n = 1) |>
ungroup() |>
select(1 : 3, contains("gdp"))You need to be very careful making an optimization like this!
Joins
This interactive plot uses the tooltip to show the airport code for the destination:
library(plotly)
p <- ggplot(fl_dest) +
geom_point(aes(x = n,
y = ave_arr_delay,
text = dest)) +
thm
## Warning in geom_point(aes(x = n, y = ave_arr_delay, text = dest)): Ignoring
## unknown aesthetics: text
ggplotly(p, tooltip = "text")A nicer approach would be to show the airport name.
The airports table provides this information:
head(airports, 2)
## # A tibble: 2 × 8
## faa name lat lon alt tz dst tzone
## <chr> <chr> <dbl> <dbl> <dbl> <dbl> <chr> <chr>
## 1 04G Lansdowne Airport 41.1 -80.6 1044 -5 A America/New…
## 2 06A Moton Field Municipal Airport 32.5 -85.7 264 -6 A America/Chi…We need to get this information into the fl_dest data frame.
As another example, we might want to explore whether aircraft age is related to delays.
The planes table provides the year of manufacture:
head(planes, 2)
## # A tibble: 2 × 9
## tailnum year type manufacturer model engines seats speed engine
## <chr> <int> <chr> <chr> <chr> <int> <int> <int> <chr>
## 1 N10156 2004 Fixed wing multi … EMBRAER EMB-… 2 55 NA Turbo…
## 2 N102UW 1998 Fixed wing multi … AIRBUS INDU… A320… 2 182 NA Turbo…Again, we need to merge this information with the data in the flights table.
One way to do this is with a join operation.
Joins are a common operation in data bases.
Murell’s Data Technologies book describes data bases in Section 5.6 and data base queries using SQL in Chapter 7.
R for Data Science describes relational data in Chapter 13.
dplyr distinguishes between two kinds of joins:
mutating joins;
filtering joins.
These animated explanations may be helpful.
Mutating Joins
Mutating joins combine variables and rows from two tables by matching rows on keys.
A primary key is a variable, or combination of variables, in a table that uniquely identify the rows of the table.
A foreign key is a variable, or combination of variables, that uniquely identify a row in some table (usually, but not necessarily, a different table).
The simplest join operation is an inner join: Only rows for which the key appears in both tables are retained.
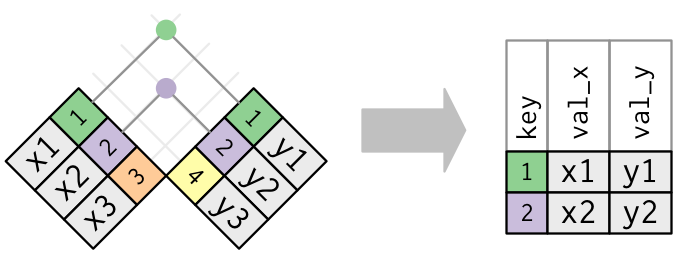
Outer joins retain some other rows with NA values for new variables:
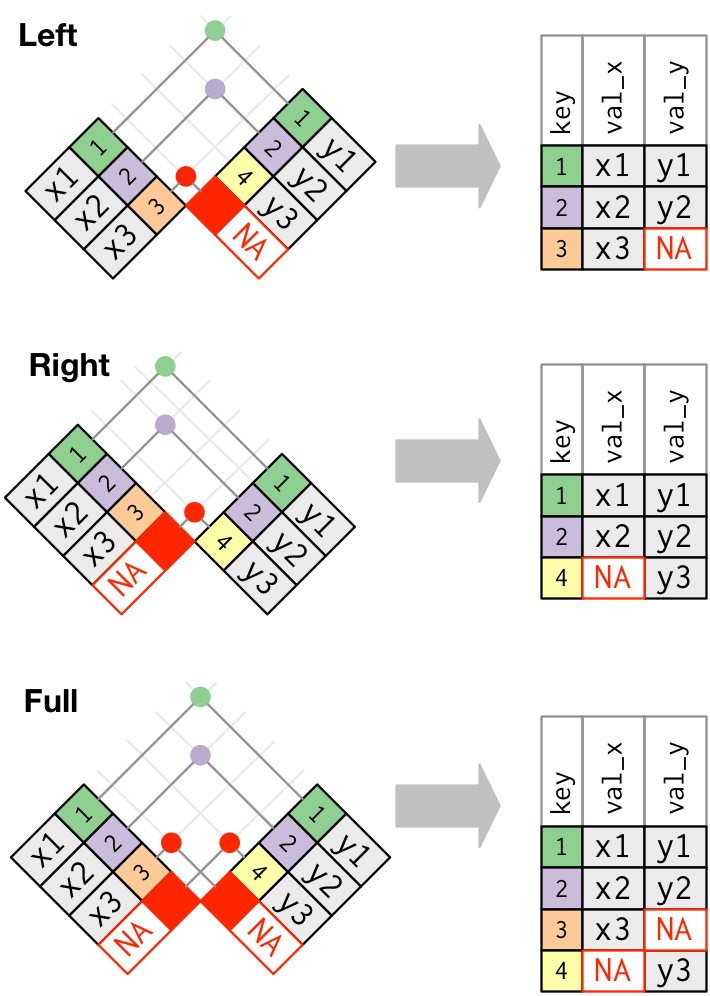
Inner and left joins are the most common.
The key of one of the tables should usually be a primary key that uniquely identifies the table’s rows.
For a left join, the right table key should usually use a primary key.
It is usually a good idea to make sure a primary key candidate really does uniquely identify rows.
For the planes table the tailnum variable is a primary key;
count(planes, tailnum) |> filter(n > 1)
## # A tibble: 0 × 2
## # ℹ 2 variables: tailnum <chr>, n <int>For the flights data a primary key needs to use multiple variables.
A reasonable guess is to use the date, carrier and flight number, but this isn’t quite enough:
count(flights, year, month, day, flight, carrier) |>
filter(n > 1) |>
head(2)
## # A tibble: 2 × 6
## year month day flight carrier n
## <int> <int> <int> <int> <chr> <int>
## 1 2013 6 8 2269 WN 2
## 2 2013 6 15 2269 WN 2Adding the origin airport code does produce a primary key:
count(flights, year, month, day, flight, carrier, origin) |>
filter(n > 1)
## # A tibble: 0 × 7
## # ℹ 7 variables: year <int>, month <int>, day <int>, flight <int>,
## # carrier <chr>, origin <chr>, n <int>To relate delays to aircraft age we can start with summaries by tailnum and year (year would be needed if we had data for more than 2013):
fl <- filter(flights, ! is.na(dep_time), ! is.na(arr_time)) |>
group_by(year, tailnum) |>
summarize(ave_dep_delay = mean(dep_delay, na.rm = TRUE),
ave_arr_delay = mean(arr_delay, na.rm = TRUE),
n = n()) |>
ungroup()head(fl)
## # A tibble: 6 × 5
## year tailnum ave_dep_delay ave_arr_delay n
## <int> <chr> <dbl> <dbl> <int>
## 1 2013 D942DN 31.5 31.5 4
## 2 2013 N0EGMQ 8.49 9.98 354
## 3 2013 N10156 17.8 12.7 146
## 4 2013 N102UW 8 2.94 48
## 5 2013 N103US -3.20 -6.93 46
## 6 2013 N104UW 10.1 1.80 46A reduced planes table with year renamed to avoid a conflict:
pl <- select(planes, tailnum, plane_year = year)pl
## # A tibble: 3,322 × 2
## tailnum plane_year
## <chr> <int>
## 1 N10156 2004
## 2 N102UW 1998
## 3 N103US 1999
## 4 N104UW 1999
## 5 N10575 2002
## 6 N105UW 1999
## 7 N107US 1999
## 8 N108UW 1999
## 9 N109UW 1999
## 10 N110UW 1999
## # ℹ 3,312 more rowsNow
use a left join with
tailnumas the key to bring theplane_yearvalue into the summary tableand then compute the plane age:
fl_age <- left_join(fl, pl, "tailnum") |>
mutate(plane_age = year - plane_year)
fl_age
## # A tibble: 4,037 × 7
## year tailnum ave_dep_delay ave_arr_delay n plane_year plane_age
## <int> <chr> <dbl> <dbl> <int> <int> <int>
## 1 2013 D942DN 31.5 31.5 4 NA NA
## 2 2013 N0EGMQ 8.49 9.98 354 NA NA
## 3 2013 N10156 17.8 12.7 146 2004 9
## 4 2013 N102UW 8 2.94 48 1998 15
## 5 2013 N103US -3.20 -6.93 46 1999 14
## 6 2013 N104UW 10.1 1.80 46 1999 14
## 7 2013 N10575 22.3 20.7 271 2002 11
## 8 2013 N105UW 2.58 -0.267 45 1999 14
## 9 2013 N107US -0.463 -5.73 41 1999 14
## 10 2013 N108UW 4.22 -1.25 60 1999 14
## # ℹ 4,027 more rowsAn initial plot:
ggplot(fl_age,
aes(x = plane_age,
y = ave_dep_delay)) +
geom_point() +
thm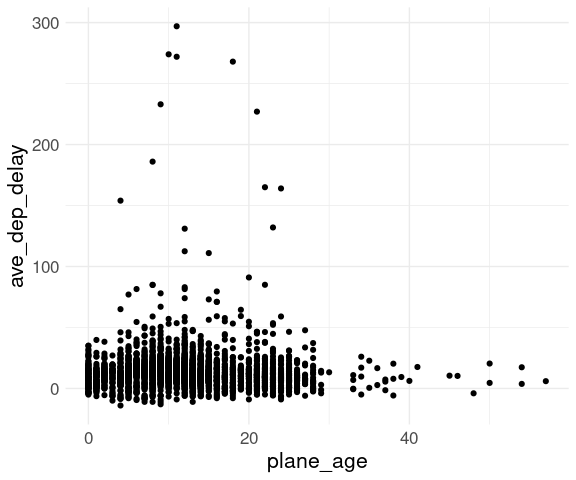
Jittering and adjusting the point size may help:
ggplot(fl_age,
aes(x = plane_age,
y = ave_dep_delay)) +
geom_point(position = "jitter",
size = 0.1) +
thm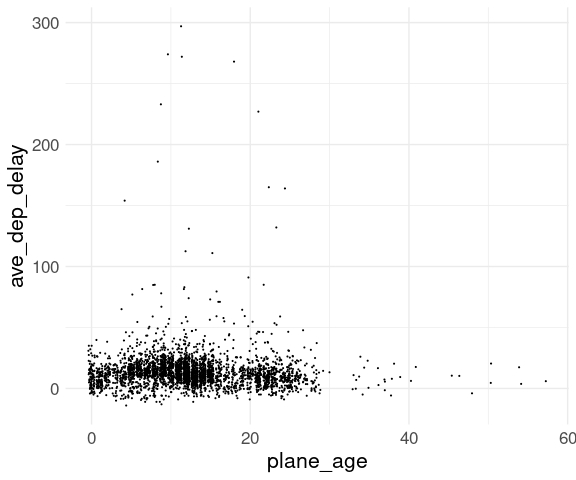
Adding a smooth and adjusting the ranges may also help:
ggplot(fl_age,
aes(x = plane_age,
y = ave_dep_delay)) +
geom_point(position = "jitter",
size = 0.1) +
geom_smooth() +
xlim(0, 30) +
ylim(c(-20, 80)) +
thm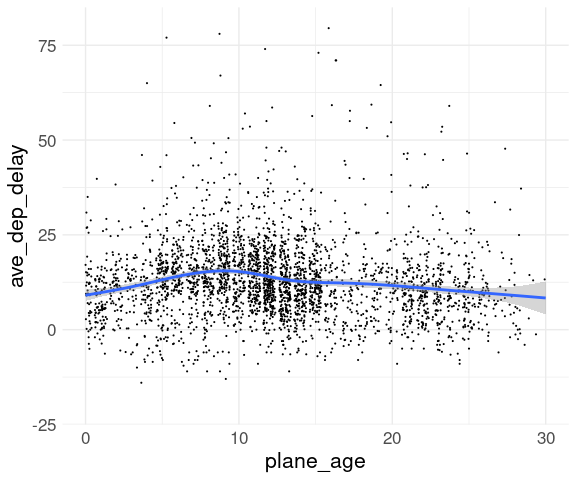
Another option is to compute means or medians within years:
flm <- group_by(fl_age, plane_age) |>
summarize(ave_dep_delay =
mean(ave_dep_delay,
na.rm = TRUE)) |>
ungroup()
ggplot(flm,
aes(x = plane_age,
y = ave_dep_delay)) +
geom_point(na.rm = TRUE) +
thm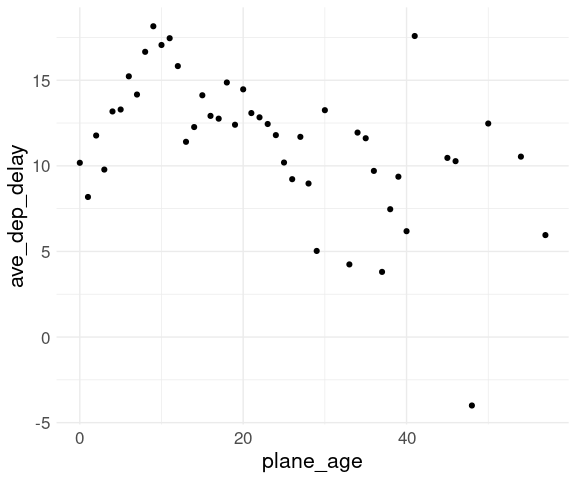
Both the smooth and the means show a modest increase over the first 10 years.
A left join can also be used to add the airport name from the airport table to the fl_dest table.
The primary key in the airport table is the three-letter airport code in the faa variable:
head(airports, 2)
## # A tibble: 2 × 8
## faa name lat lon alt tz dst tzone
## <chr> <chr> <dbl> <dbl> <dbl> <dbl> <chr> <chr>
## 1 04G Lansdowne Airport 41.1 -80.6 1044 -5 A America/New…
## 2 06A Moton Field Municipal Airport 32.5 -85.7 264 -6 A America/Chi…The foreign key in the fl_dest table to match is the dest variable:
head(fl_dest, 2)
## # A tibble: 2 × 6
## dest ave_dep_delay ave_arr_delay max_dep_delay max_arr_delay n
## <chr> <dbl> <dbl> <dbl> <dbl> <int>
## 1 ABQ 13.7 4.38 142 153 254
## 2 ACK 6.46 4.85 219 221 265The left_join function allows this link to be made by specifying the key as c("dest" = "faa"):
fl_dest <- left_join(fl_dest,
select(airports, faa, dest_name = name),
c("dest" = "faa"))
select(fl_dest, dest_name, everything())
## # A tibble: 105 × 7
## dest_name dest ave_dep_delay ave_arr_delay max_dep_delay max_arr_delay n
## <chr> <chr> <dbl> <dbl> <dbl> <dbl> <int>
## 1 Albuquer… ABQ 13.7 4.38 142 153 254
## 2 Nantucke… ACK 6.46 4.85 219 221 265
## 3 Albany I… ALB 23.6 14.4 323 328 439
## 4 Ted Stev… ANC 12.9 -2.5 75 39 8
## 5 Hartsfie… ATL 12.5 11.3 898 895 17215
## 6 Austin B… AUS 13.0 6.02 351 349 2439
## 7 Ashevill… AVL 8.19 8.00 222 228 275
## 8 Bradley … BDL 17.7 7.05 252 266 443
## 9 Bangor I… BGR 19.5 8.03 248 238 375
## 10 Birmingh… BHM 29.7 16.9 325 291 297
## # ℹ 95 more rowsNow a tooltip can provide a more accessible label:
p <- ggplot(fl_dest) +
geom_point(aes(x = n,
y = ave_arr_delay,
text = paste(dest,
dest_name,
sep = ": "))) +
thm
ggplotly(p, tooltip = "text")Filtering Joins
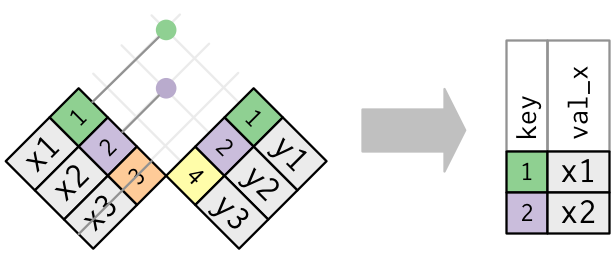
Filtering joins only affect rows; they do not add variables.
semi_join(x, y)returns all rows inxwith a match iny:
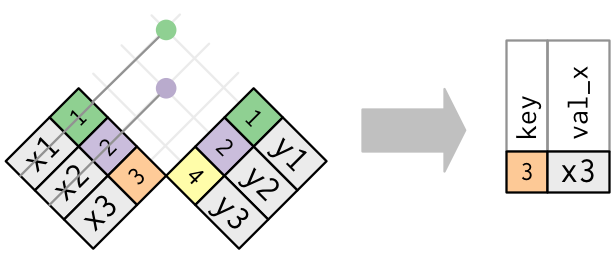
anti_join(x, y)returns all rows inxwithout a match iny:
semi_join is useful for matching filtered results back to a table.
The plot of delay against age showed some surprisingly old aircraft:
top_age <- slice_max(fl_age, plane_age, n = 2)
top_age
## # A tibble: 3 × 7
## year tailnum ave_dep_delay ave_arr_delay n plane_year plane_age
## <int> <chr> <dbl> <dbl> <int> <int> <int>
## 1 2013 N381AA 5.95 3.5 22 1956 57
## 2 2013 N201AA 17.4 14.1 51 1959 54
## 3 2013 N567AA 3.72 -4.30 61 1959 54We can identify these aircraft with a semi-join:
semi_join(planes, top_age, "tailnum")
## # A tibble: 3 × 9
## tailnum year type manufacturer model engines seats speed engine
## <chr> <int> <chr> <chr> <chr> <int> <int> <int> <chr>
## 1 N201AA 1959 Fixed wing single… CESSNA 150 1 2 90 Recip…
## 2 N381AA 1956 Fixed wing multi … DOUGLAS DC-7… 4 102 232 Recip…
## 3 N567AA 1959 Fixed wing single… DEHAVILLAND OTTE… 1 16 95 Recip…The corresponding flights can also be picked out with a semi-join:
top_age_flights <- semi_join(flights, top_age, "tailnum")
head(top_age_flights, 2)
## # A tibble: 2 × 19
## year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time
## <int> <int> <int> <int> <int> <dbl> <int> <int>
## 1 2013 1 3 909 700 129 1103 850
## 2 2013 1 3 NA 920 NA NA 1245
## # ℹ 11 more variables: arr_delay <dbl>, carrier <chr>, flight <int>,
## # tailnum <chr>, origin <chr>, dest <chr>, air_time <dbl>, distance <dbl>,
## # hour <dbl>, minute <dbl>, time_hour <dttm>Looking at the carriers or destinations might help:
select(top_age_flights, carrier, dest, everything()) |> head(4)
## # A tibble: 4 × 19
## carrier dest year month day dep_time sched_dep_time dep_delay arr_time
## <chr> <chr> <int> <int> <int> <int> <int> <dbl> <int>
## 1 AA ORD 2013 1 3 909 700 129 1103
## 2 AA DFW 2013 1 3 NA 920 NA NA
## 3 AA DFW 2013 1 9 1423 1430 -7 1727
## 4 AA DFW 2013 1 10 1238 1240 -2 1525
## # ℹ 10 more variables: sched_arr_time <int>, arr_delay <dbl>, flight <int>,
## # tailnum <chr>, origin <chr>, air_time <dbl>, distance <dbl>, hour <dbl>,
## # minute <dbl>, time_hour <dttm>The carrier summary:
count(top_age_flights, carrier)
## # A tibble: 1 × 2
## carrier n
## <chr> <int>
## 1 AA 139A check of the documentation for planes shows that American and one other airline report fleet numbers rather than tail numbers.
Anti-joins are useful for detecting join problems.
For example, many flights have tail numbers that do not match entries in planes:
fl <- anti_join(flights, planes, "tailnum")
nrow(fl)
## [1] 52606count(fl, tailnum)
## # A tibble: 722 × 2
## tailnum n
## <chr> <int>
## 1 D942DN 4
## 2 N0EGMQ 371
## 3 N14628 1
## 4 N149AT 22
## 5 N16632 11
## 6 N17627 2
## 7 N1EAMQ 210
## 8 N200AA 34
## 9 N24633 8
## 10 N261AV 6
## # ℹ 712 more rowsJoin Issues
Make sure a primary key you want to use has no missing values.
Make sure a primary key you want to use really uniquely identifies a row.
Make sure every value of your foreign key you want to use to link to another table matches a row in the linked table (
anti_joinis a good way to do this).Check that your foreign key has no missing values.
Data Base Interface
Current data on airline flights in the US is available from the Bureau of Transportation Statistics at
http://www.transtats.bts.gov/DL_SelectFields.asp?Table_ID=236
The data are available in one compressed CSV file per month.
Loaded into R, one month of data requires about 300 Mb of memory.
Data for 1987–2008 was compiled for a DataExpo competition of the American Statistical Association held in 2009.
The data are available on the Linux systems in the directory /group/statsoft/data/DataExpo09 as compressed CSV files and as a SQLite data base.
Loading the data into R would require about 70 Gb of memory, which is not practical.
Using the data base is a good alternative.
SQLite is a simple relational data base that works with data stored on a local disk.
More sophisticated data bases allow data to be stored on multiple machines or in the cloud
These data bases are usually accessed by connecting to a server, which usually involves an authentication process.
For SQLite the connection is set up by linking to a file.
The RSQLite package provides an interface to SQLite.
The DBI package provides a high level interface for using data bases.
dbConnectcreates a generic data base connection object.dbListTablesreturns a vector with the names of the available tables.dbDisconnectcloses a data base connection.
Analogous packages exist for Python and Perl.
The first step is to connect to the data base.
library(dplyr)
library(DBI)
library(RSQLite)
db <- dbConnect(SQLite(), "/group/statsoft/data/DataExpo09/ontime.sqlite3")There is only one table:
dbListTables(db)
## [1] "ontime"tbl creates a tibble-like object for a data base table.
fl <- tbl(db, "ontime")This object is not a regular tibble, but many tibble operations work on it.
To find the variables in the table:
tbl_vars(fl)
## [1] "Year" "Month" "DayofMonth"
## [4] "DayOfWeek" "DepTime" "CRSDepTime"
## [7] "ArrTime" "CRSArrTime" "UniqueCarrier"
## [10] "FlightNum" "TailNum" "ActualElapsedTime"
## [13] "CRSElapsedTime" "AirTime" "ArrDelay"
## [16] "DepDelay" "Origin" "Dest"
## [19] "Distance" "TaxiIn" "TaxiOut"
## [22] "Cancelled" "CancellationCode" "Diverted"
## [25] "CarrierDelay" "WeatherDelay" "NASDelay"
## [28] "SecurityDelay" "LateAircraftDelay"Many dplyr operations are supported:
sm <- summarize(fl, last_year = max(Year, na.rm = TRUE), n = n())This computation is fast because it does not actually access the data base.
An operation that needs the data, such as printing the result, will access the data base and take time:
sm
## # Source: lazy query [?? x 2]
## # Database: sqlite 3.37.2
## # [/mnt/nfs/clasnetappvm/group/statsoft/data/DataExpo09/ontime.sqlite3]
## last_year n
## <int> <int>
## 1 2008 118914458The sm object essentially contains a SQL query, which can be examined using sql_render from the dbplyr package:
dbplyr::sql_render(sm)
## <SQL> SELECT MAX(`Year`) AS `last_year`, COUNT(*) AS `n`
## FROM `ontime`Flights to CID in 2008:
fl_cid <- filter(fl, Year == 2008, Dest == "CID")
dbplyr::sql_render(fl_cid)
## <SQL> SELECT *
## FROM `ontime`
## WHERE ((`Year` = 2008.0) AND (`Dest` = 'CID'))Number of flights to CID in 2008:
sm <- summarize(fl_cid, n = n())
dbplyr::sql_render(sm)
## <SQL> SELECT COUNT(*) AS `n`
## FROM `ontime`
## WHERE ((`Year` = 2008.0) AND (`Dest` = 'CID'))sm
## # Source: lazy query [?? x 1]
## # Database: sqlite 3.37.2
## # [/mnt/nfs/clasnetappvm/group/statsoft/data/DataExpo09/ontime.sqlite3]
## n
## <int>
## 1 2961Breakdown by where the flights originated:
sm <- group_by(fl_cid, Origin) |>
summarize(n = n()) |>
ungroup()
dbplyr::sql_render(sm)
## <SQL> SELECT `Origin`, COUNT(*) AS `n`
## FROM `ontime`
## WHERE ((`Year` = 2008.0) AND (`Dest` = 'CID'))
## GROUP BY `Origin`sm
## # Source: lazy query [?? x 2]
## # Database: sqlite 3.37.2
## # [/mnt/nfs/clasnetappvm/group/statsoft/data/DataExpo09/ontime.sqlite3]
## Origin n
## <chr> <int>
## 1 ATL 119
## 2 BNA 1
## 3 CVG 292
## 4 DEN 230
## 5 DFW 519
## 6 DTW 345
## 7 MSP 241
## 8 ORD 1214Many base R functions can be converted to SQL:
sm <- group_by(fl_cid, Origin) |>
summarize(ave_arr_del_pos = mean(pmax(ArrDelay, 0), na.rm = TRUE)) |>
ungroup()
dbplyr::sql_render(sm)
## <SQL> SELECT `Origin`, AVG(MAX(`ArrDelay`, 0.0)) AS `ave_arr_del_pos`
## FROM `ontime`
## WHERE ((`Year` = 2008.0) AND (`Dest` = 'CID'))
## GROUP BY `Origin`sm_cid <- sm
sm
## # Source: lazy query [?? x 2]
## # Database: sqlite 3.37.2
## # [/mnt/nfs/clasnetappvm/group/statsoft/data/DataExpo09/ontime.sqlite3]
## Origin ave_arr_del_pos
## <chr> <dbl>
## 1 ATL 17.4
## 2 BNA 18
## 3 CVG 12.4
## 4 DEN 12.1
## 5 DFW 11.3
## 6 DTW 15.0
## 7 MSP 20.1
## 8 ORD 21.6mutate() can also be used:
fl_cid_gain <- mutate(fl_cid, Gain = DepDelay - ArrDelay)
dbplyr::sql_render(fl_cid_gain)
## <SQL> SELECT `Year`, `Month`, `DayofMonth`, `DayOfWeek`, `DepTime`, `CRSDepTime`, `ArrTime`, `CRSArrTime`, `UniqueCarrier`, `FlightNum`, `TailNum`, `ActualElapsedTime`, `CRSElapsedTime`, `AirTime`, `ArrDelay`, `DepDelay`, `Origin`, `Dest`, `Distance`, `TaxiIn`, `TaxiOut`, `Cancelled`, `CancellationCode`, `Diverted`, `CarrierDelay`, `WeatherDelay`, `NASDelay`, `SecurityDelay`, `LateAircraftDelay`, `DepDelay` - `ArrDelay` AS `Gain`
## FROM `ontime`
## WHERE ((`Year` = 2008.0) AND (`Dest` = 'CID'))group_by(fl_cid_gain, Origin) |>
summarize(mean_gain = mean(Gain)) |>
ungroup()
## # Source: lazy query [?? x 2]
## # Database: sqlite 3.37.2
## # [/mnt/nfs/clasnetappvm/group/statsoft/data/DataExpo09/ontime.sqlite3]
## Origin mean_gain
## <chr> <dbl>
## 1 ATL 8.24
## 2 BNA -26
## 3 CVG -1.60
## 4 DEN 0.278
## 5 DFW 4.12
## 6 DTW 3.04
## 7 MSP -0.357
## 8 ORD -1.36Once a filtered result is small enough it can be brought over as a regular tibble using
tb <- collect(fl_cid)There are some limitations to operations that can be done on tables in data bases.
Defining and using a function works for a tibble:
pos_mean <- function(x) mean(pmax(x, 0))
group_by(tb, Origin) |>
summarize(ave_arr_del_pos = pos_mean(ArrDelay)) |>
ungroup()But this fails:
group_by(fl_cid, Origin) |>
summarize(ave_arr_del_pos = pos_mean(ArrDelay)) |>
ungroup()
## Error: no such function: pos_meanSome operations may require some modifications.
Recomputing sm_cid:
fl <- tbl(db, "ontime")
fl_cid <- filter(fl, Year == 2008, Dest == "CID")
sm_cid <- group_by(fl_cid, Origin) |>
summarize(ave_arr_del_pos = mean(pmax(ArrDelay, 0), na.rm = TRUE)) |>
ungroup()Joining with a local table does not work:
ap <- select(nycflights13::airports, faa, name)
sm_cid <- left_join(sm_cid, ap, c(Origin = "faa"))
## Error in `auto_copy()`:
## ! `x` and `y` must share the same src.
## ℹ set `copy` = TRUE (may be slow).But this does work:
sm_cid <- left_join(sm_cid, ap, c(Origin = "faa"), copy = TRUE)This copies the ap table to the data base:
dbListTables(db)
## [1] "dbplyr_001" "ontime" "sqlite_stat1" "sqlite_stat4"Finish by disconnecting from the data base:
dbDisconnect(db)This is not necessary for SQLite but is for other data bases, so it is good practice.
Some notes:
Building data base queries with
dplyris very efficient.Running the queries can take time.
Transferring (partial) results to R can take time.
Inadvertently asking to transfer a result that is too large could cause problems.
Other data bases may arrange to cache query results.
Most data bases, including SQLite, support creating indices that speed up certain queries at the expense of using more storage.
The dbplyr web site provides more information on using data bases with dplyr.
Some other approaches for handling larger-than-memory data:
Apache arrow and the R
arrowpackage.Polars and the
polarsandtidypolarsR packages.
Other Wrangling Tools
Some additional tools provided by tidyr:
separate(): split character variable into several variables;fill(): fill in missing values in a variable (e.g. carry forward);complete(): add missing category classes (e.g. zero counts).
We have already seen pivot_longer() and pivot_wider().
A newer alternative to separate() is to use separate_wider_position() and separate_wider_delim()
Separating Variables
A data set from the WHO on tuberculosis cases:
head(who, 2)
## # A tibble: 2 × 60
## country iso2 iso3 year new_sp_m014 new_sp_m1524 new_sp_m2534 new_sp_m3544
## <chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 Afghanis… AF AFG 1980 NA NA NA NA
## 2 Afghanis… AF AFG 1981 NA NA NA NA
## # ℹ 52 more variables: new_sp_m4554 <dbl>, new_sp_m5564 <dbl>,
## # new_sp_m65 <dbl>, new_sp_f014 <dbl>, new_sp_f1524 <dbl>,
## # new_sp_f2534 <dbl>, new_sp_f3544 <dbl>, new_sp_f4554 <dbl>,
## # new_sp_f5564 <dbl>, new_sp_f65 <dbl>, new_sn_m014 <dbl>,
## # new_sn_m1524 <dbl>, new_sn_m2534 <dbl>, new_sn_m3544 <dbl>,
## # new_sn_m4554 <dbl>, new_sn_m5564 <dbl>, new_sn_m65 <dbl>,
## # new_sn_f014 <dbl>, new_sn_f1524 <dbl>, new_sn_f2534 <dbl>, …Some of the remaining variable names:
names(who) |> tail(20)
## [1] "new_ep_f1524" "new_ep_f2534" "new_ep_f3544" "new_ep_f4554" "new_ep_f5564"
## [6] "new_ep_f65" "newrel_m014" "newrel_m1524" "newrel_m2534" "newrel_m3544"
## [11] "newrel_m4554" "newrel_m5564" "newrel_m65" "newrel_f014" "newrel_f1524"
## [16] "newrel_f2534" "newrel_f3544" "newrel_f4554" "newrel_f5564" "newrel_f65"The variable new_sp_m2534, for example, represents the number of cases for
- type
sp(positive pulmonary smear); - sex
m; - age between 25 and 34.
It would be better (tidier) to have separate variables for
- count
- type
- sex
- lower bound on age bracket
- upper bound on age bracket
A pipeline to clean this up involves pivoting to long form and several separate and mutate steps.
The original data frame:
who
## # A tibble: 7,240 × 60
## country iso2 iso3 year new_sp_m014 new_sp_m1524 new_sp_m2534 new_sp_m3544
## <chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 Afghani… AF AFG 1980 NA NA NA NA
## 2 Afghani… AF AFG 1981 NA NA NA NA
## 3 Afghani… AF AFG 1982 NA NA NA NA
## 4 Afghani… AF AFG 1983 NA NA NA NA
## 5 Afghani… AF AFG 1984 NA NA NA NA
## 6 Afghani… AF AFG 1985 NA NA NA NA
## 7 Afghani… AF AFG 1986 NA NA NA NA
## 8 Afghani… AF AFG 1987 NA NA NA NA
## 9 Afghani… AF AFG 1988 NA NA NA NA
## 10 Afghani… AF AFG 1989 NA NA NA NA
## # ℹ 7,230 more rows
## # ℹ 52 more variables: new_sp_m4554 <dbl>, new_sp_m5564 <dbl>,
## # new_sp_m65 <dbl>, new_sp_f014 <dbl>, new_sp_f1524 <dbl>,
## # new_sp_f2534 <dbl>, new_sp_f3544 <dbl>, new_sp_f4554 <dbl>,
## # new_sp_f5564 <dbl>, new_sp_f65 <dbl>, new_sn_m014 <dbl>,
## # new_sn_m1524 <dbl>, new_sn_m2534 <dbl>, new_sn_m3544 <dbl>,
## # new_sn_m4554 <dbl>, new_sn_m5564 <dbl>, new_sn_m65 <dbl>, …First pivot to longer form:
who_clean <-
pivot_longer(who,
new_sp_m014 : newrel_f65,
names_to = "key",
values_to = "count")who_clean
## # A tibble: 405,440 × 6
## country iso2 iso3 year key count
## <chr> <chr> <chr> <dbl> <chr> <dbl>
## 1 Afghanistan AF AFG 1980 new_sp_m014 NA
## 2 Afghanistan AF AFG 1980 new_sp_m1524 NA
## 3 Afghanistan AF AFG 1980 new_sp_m2534 NA
## 4 Afghanistan AF AFG 1980 new_sp_m3544 NA
## 5 Afghanistan AF AFG 1980 new_sp_m4554 NA
## 6 Afghanistan AF AFG 1980 new_sp_m5564 NA
## 7 Afghanistan AF AFG 1980 new_sp_m65 NA
## 8 Afghanistan AF AFG 1980 new_sp_f014 NA
## 9 Afghanistan AF AFG 1980 new_sp_f1524 NA
## 10 Afghanistan AF AFG 1980 new_sp_f2534 NA
## # ℹ 405,430 more rowsRemove "new" or "new_" prefix:
who_clean <-
pivot_longer(who,
new_sp_m014 : newrel_f65,
names_to = "key",
values_to = "count") |>
mutate(key = sub("new_?", "", key))who_clean
## # A tibble: 405,440 × 6
## country iso2 iso3 year key count
## <chr> <chr> <chr> <dbl> <chr> <dbl>
## 1 Afghanistan AF AFG 1980 sp_m014 NA
## 2 Afghanistan AF AFG 1980 sp_m1524 NA
## 3 Afghanistan AF AFG 1980 sp_m2534 NA
## 4 Afghanistan AF AFG 1980 sp_m3544 NA
## 5 Afghanistan AF AFG 1980 sp_m4554 NA
## 6 Afghanistan AF AFG 1980 sp_m5564 NA
## 7 Afghanistan AF AFG 1980 sp_m65 NA
## 8 Afghanistan AF AFG 1980 sp_f014 NA
## 9 Afghanistan AF AFG 1980 sp_f1524 NA
## 10 Afghanistan AF AFG 1980 sp_f2534 NA
## # ℹ 405,430 more rowsSeparate key into type and sexage:
who_clean <-
pivot_longer(who,
new_sp_m014 : newrel_f65,
names_to = "key",
values_to = "count") |>
mutate(key = sub("new_?", "", key)) |>
separate(key, c("type", "sexage"))who_clean
## # A tibble: 405,440 × 7
## country iso2 iso3 year type sexage count
## <chr> <chr> <chr> <dbl> <chr> <chr> <dbl>
## 1 Afghanistan AF AFG 1980 sp m014 NA
## 2 Afghanistan AF AFG 1980 sp m1524 NA
## 3 Afghanistan AF AFG 1980 sp m2534 NA
## 4 Afghanistan AF AFG 1980 sp m3544 NA
## 5 Afghanistan AF AFG 1980 sp m4554 NA
## 6 Afghanistan AF AFG 1980 sp m5564 NA
## 7 Afghanistan AF AFG 1980 sp m65 NA
## 8 Afghanistan AF AFG 1980 sp f014 NA
## 9 Afghanistan AF AFG 1980 sp f1524 NA
## 10 Afghanistan AF AFG 1980 sp f2534 NA
## # ℹ 405,430 more rowsSeparate sexage into sex and age:
who_clean <-
pivot_longer(who,
new_sp_m014 : newrel_f65,
names_to = "key",
values_to = "count") |>
mutate(key = sub("new_?", "", key)) |>
separate(key, c("type", "sexage")) |>
separate(sexage, c("sex", "age"),
sep = 1)who_clean
## # A tibble: 405,440 × 8
## country iso2 iso3 year type sex age count
## <chr> <chr> <chr> <dbl> <chr> <chr> <chr> <dbl>
## 1 Afghanistan AF AFG 1980 sp m 014 NA
## 2 Afghanistan AF AFG 1980 sp m 1524 NA
## 3 Afghanistan AF AFG 1980 sp m 2534 NA
## 4 Afghanistan AF AFG 1980 sp m 3544 NA
## 5 Afghanistan AF AFG 1980 sp m 4554 NA
## 6 Afghanistan AF AFG 1980 sp m 5564 NA
## 7 Afghanistan AF AFG 1980 sp m 65 NA
## 8 Afghanistan AF AFG 1980 sp f 014 NA
## 9 Afghanistan AF AFG 1980 sp f 1524 NA
## 10 Afghanistan AF AFG 1980 sp f 2534 NA
## # ℹ 405,430 more rowsFix up age categories:
who_clean <-
pivot_longer(who,
new_sp_m014 : newrel_f65,
names_to = "key",
values_to = "count") |>
mutate(key = sub("new_?", "", key)) |>
separate(key, c("type", "sexage")) |>
separate(sexage, c("sex", "age"),
sep = 1) |>
mutate(age = sub("014", "0014", age)) |>
mutate(age = sub("65", "65Inf", age))who_clean
## # A tibble: 405,440 × 8
## country iso2 iso3 year type sex age count
## <chr> <chr> <chr> <dbl> <chr> <chr> <chr> <dbl>
## 1 Afghanistan AF AFG 1980 sp m 0014 NA
## 2 Afghanistan AF AFG 1980 sp m 1524 NA
## 3 Afghanistan AF AFG 1980 sp m 2534 NA
## 4 Afghanistan AF AFG 1980 sp m 3544 NA
## 5 Afghanistan AF AFG 1980 sp m 4554 NA
## 6 Afghanistan AF AFG 1980 sp m 5564 NA
## 7 Afghanistan AF AFG 1980 sp m 65Inf NA
## 8 Afghanistan AF AFG 1980 sp f 0014 NA
## 9 Afghanistan AF AFG 1980 sp f 1524 NA
## 10 Afghanistan AF AFG 1980 sp f 2534 NA
## # ℹ 405,430 more rowsFinally, split age into age_lo and age-hi:
who_clean <-
pivot_longer(who,
new_sp_m014 : newrel_f65,
names_to = "key",
values_to = "count") |>
mutate(key = sub("new_?", "", key)) |>
separate(key, c("type", "sexage")) |>
separate(sexage, c("sex", "age"),
sep = 1) |>
mutate(age = sub("014", "0014", age)) |>
mutate(age = sub("65", "65Inf", age)) |>
separate(age, c("age_lo", "age_hi"),
sep = 2,
convert = TRUE)who_clean
## # A tibble: 405,440 × 9
## country iso2 iso3 year type sex age_lo age_hi count
## <chr> <chr> <chr> <dbl> <chr> <chr> <int> <dbl> <dbl>
## 1 Afghanistan AF AFG 1980 sp m 0 14 NA
## 2 Afghanistan AF AFG 1980 sp m 15 24 NA
## 3 Afghanistan AF AFG 1980 sp m 25 34 NA
## 4 Afghanistan AF AFG 1980 sp m 35 44 NA
## 5 Afghanistan AF AFG 1980 sp m 45 54 NA
## 6 Afghanistan AF AFG 1980 sp m 55 64 NA
## 7 Afghanistan AF AFG 1980 sp m 65 Inf NA
## 8 Afghanistan AF AFG 1980 sp f 0 14 NA
## 9 Afghanistan AF AFG 1980 sp f 15 24 NA
## 10 Afghanistan AF AFG 1980 sp f 25 34 NA
## # ℹ 405,430 more rowsA plot of total numbers of cases for a few countries over time:
filter(who_clean,
iso3 %in% c("CAN", "DEU", "GBR", "USA")) |>
group_by(year, iso3) |>
summarize(count = sum(count, na.rm = TRUE)) |>
ungroup() |>
ggplot() +
geom_line(aes(x = year,
y = count,
color = iso3)) +
thm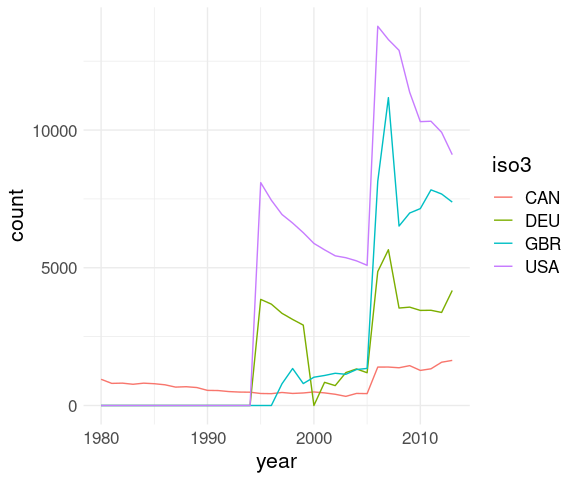
Filling In Omitted Values
The Refugee Processing Center provides information from the United States Refugee Admissions Program (USRAP), including arrivals by state and nationality.
Three files are available locally:
data from early January 2017;
data from early April 2017.
data from early March 2018.
These are Excel spread sheets.
Reading in the third file, with data for October 2017 through February 2018:
ref_url <-
"http://homepage.stat.uiowa.edu/~luke/data/Arrivals-2018-03-05.xls"
ref_file <- "Arrivals-2018-03-05.xls"
if (! file.exists(ref_file))
download.file(ref_url, ref_file)
library(readxl)
ref <- read_excel(ref_file, skip = 16) ## skip header
ref <- head(ref, -2) ## drop last two rows
names(ref)[1] <- "Destination"The Destination column needs filling in (this is common in spread sheet data):
ref
## # A tibble: 500 × 18
## Destination Nationality Oct Nov Dec Jan Feb Mar Apr May Jun
## <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 Alabama <NA> 0 0 3 1 4 0 0 0 0
## 2 <NA> Dem. Rep. … 0 0 3 0 4 0 0 0 0
## 3 <NA> El Salvador 0 0 0 1 0 0 0 0 0
## 4 Alaska <NA> 9 2 6 0 0 0 0 0 0
## 5 <NA> Russia 0 0 6 0 0 0 0 0 0
## 6 <NA> Ukraine 9 2 0 0 0 0 0 0 0
## 7 Arizona <NA> 26 47 71 73 110 0 0 0 0
## 8 <NA> Afghanistan 0 1 14 10 17 0 0 0 0
## 9 <NA> Algeria 0 0 0 0 1 0 0 0 0
## 10 <NA> Bhutan 2 3 8 2 0 0 0 0 0
## # ℹ 490 more rows
## # ℹ 7 more variables: Jul <dbl>, Aug <dbl>, Sep <dbl>, Cases <dbl>, Inds <dbl>,
## # State <dbl>, U.S. <dbl>Fill down the Destination:
ref_clean <-
fill(ref, Destination)ref_clean
## # A tibble: 500 × 18
## Destination Nationality Oct Nov Dec Jan Feb Mar Apr May Jun
## <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 Alabama <NA> 0 0 3 1 4 0 0 0 0
## 2 Alabama Dem. Rep. … 0 0 3 0 4 0 0 0 0
## 3 Alabama El Salvador 0 0 0 1 0 0 0 0 0
## 4 Alaska <NA> 9 2 6 0 0 0 0 0 0
## 5 Alaska Russia 0 0 6 0 0 0 0 0 0
## 6 Alaska Ukraine 9 2 0 0 0 0 0 0 0
## 7 Arizona <NA> 26 47 71 73 110 0 0 0 0
## 8 Arizona Afghanistan 0 1 14 10 17 0 0 0 0
## 9 Arizona Algeria 0 0 0 0 1 0 0 0 0
## 10 Arizona Bhutan 2 3 8 2 0 0 0 0 0
## # ℹ 490 more rows
## # ℹ 7 more variables: Jul <dbl>, Aug <dbl>, Sep <dbl>, Cases <dbl>, Inds <dbl>,
## # State <dbl>, U.S. <dbl>Drop the totals:
ref_clean <-
fill(ref, Destination) |>
filter(! is.na(Nationality))ref_clean
## # A tibble: 451 × 18
## Destination Nationality Oct Nov Dec Jan Feb Mar Apr May Jun
## <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 Alabama Dem. Rep. … 0 0 3 0 4 0 0 0 0
## 2 Alabama El Salvador 0 0 0 1 0 0 0 0 0
## 3 Alaska Russia 0 0 6 0 0 0 0 0 0
## 4 Alaska Ukraine 9 2 0 0 0 0 0 0 0
## 5 Arizona Afghanistan 0 1 14 10 17 0 0 0 0
## 6 Arizona Algeria 0 0 0 0 1 0 0 0 0
## 7 Arizona Bhutan 2 3 8 2 0 0 0 0 0
## 8 Arizona Burma 6 3 7 1 9 0 0 0 0
## 9 Arizona Burundi 0 0 0 5 7 0 0 0 0
## 10 Arizona Colombia 0 0 2 0 0 0 0 0 0
## # ℹ 441 more rows
## # ℹ 7 more variables: Jul <dbl>, Aug <dbl>, Sep <dbl>, Cases <dbl>, Inds <dbl>,
## # State <dbl>, U.S. <dbl>Pivot to a tidier form with one row per month:
ref_clean <-
fill(ref, Destination) |>
filter(! is.na(Nationality)) |>
pivot_longer(
Oct : Sep,
names_to = "month",
values_to = "count")ref_clean
## # A tibble: 5,412 × 8
## Destination Nationality Cases Inds State U.S. month count
## <chr> <chr> <dbl> <dbl> <dbl> <dbl> <chr> <dbl>
## 1 Alabama Dem. Rep. Congo 2 7 0.875 0.000811 Oct 0
## 2 Alabama Dem. Rep. Congo 2 7 0.875 0.000811 Nov 0
## 3 Alabama Dem. Rep. Congo 2 7 0.875 0.000811 Dec 3
## 4 Alabama Dem. Rep. Congo 2 7 0.875 0.000811 Jan 0
## 5 Alabama Dem. Rep. Congo 2 7 0.875 0.000811 Feb 4
## 6 Alabama Dem. Rep. Congo 2 7 0.875 0.000811 Mar 0
## 7 Alabama Dem. Rep. Congo 2 7 0.875 0.000811 Apr 0
## 8 Alabama Dem. Rep. Congo 2 7 0.875 0.000811 May 0
## 9 Alabama Dem. Rep. Congo 2 7 0.875 0.000811 Jun 0
## 10 Alabama Dem. Rep. Congo 2 7 0.875 0.000811 Jul 0
## # ℹ 5,402 more rowsTop 5 destination states:
group_by(ref_clean, Destination) |>
summarize(count = sum(count)) |>
ungroup() |>
slice_max(count, n = 5)
## # A tibble: 5 × 2
## Destination count
## <chr> <dbl>
## 1 Ohio 750
## 2 Texas 602
## 3 Washington 562
## 4 California 545
## 5 New York 540Top 5 origin nationalities:
group_by(ref_clean, Nationality) |>
summarize(count = sum(count)) |>
ungroup() |>
slice_max(count, n = 5)
## # A tibble: 5 × 2
## Nationality count
## <chr> <dbl>
## 1 Dem. Rep. Congo 1968
## 2 Bhutan 1884
## 3 Burma 1240
## 4 Ukraine 1021
## 5 Eritrea 648Completing Missing Factor Level Combinations
This example is based on a blog post using data from the Significant Earthquake Database.
The data used is a selection for North America and Hawaii.
Reading in the data:
if (! file.exists("earthquakes.tsv"))
download.file("http://homepage.stat.uiowa.edu/~luke/data/earthquakes.tsv",
"earthquakes.tsv")
eq <- read.table("earthquakes.tsv",
header = TRUE,
sep = "\t")
eq <- as_tibble(eq)select(eq, STATE, everything())
## # A tibble: 254 × 47
## STATE I_D FLAG_TSUNAMI YEAR MONTH DAY HOUR MINUTE SECOND FOCAL_DEPTH
## <chr> <int> <chr> <int> <int> <int> <int> <int> <dbl> <int>
## 1 HI 6697 Tsu 1500 NA NA NA NA NA NA
## 2 MA 6013 Tsu 1668 4 13 NA NA NA NA
## 3 OR 9954 Tsu 1700 1 27 5 0 NA NA
## 4 MA 5828 Tsu 1755 11 18 9 11 35 NA
## 5 AK 5926 Tsu 1788 7 21 NA NA NA NA
## 6 AK 5927 Tsu 1788 8 6 NA NA NA NA
## 7 AK 7057 Tsu 1792 NA NA NA NA NA NA
## 8 CA 5857 Tsu 1806 3 25 8 NA NA NA
## 9 AR 1594 Tsu 1811 12 16 8 15 NA NA
## 10 AR 7058 Tsu 1811 12 16 14 15 NA NA
## # ℹ 244 more rows
## # ℹ 37 more variables: EQ_PRIMARY <dbl>, EQ_MAG_MW <dbl>, EQ_MAG_MS <dbl>,
## # EQ_MAG_MB <dbl>, EQ_MAG_ML <dbl>, EQ_MAG_MFA <dbl>, EQ_MAG_UNK <dbl>,
## # INTENSITY <int>, COUNTRY <chr>, LOCATION_NAME <chr>, LATITUDE <dbl>,
## # LONGITUDE <dbl>, REGION_CODE <int>, DEATHS <int>, DEATHS_DESCRIPTION <int>,
## # MISSING <lgl>, MISSING_DESCRIPTION <lgl>, INJURIES <int>,
## # INJURIES_DESCRIPTION <int>, DAMAGE_MILLIONS_DOLLARS <dbl>, …Some STATE entries are blank, but the location name identifies the state:
filter(eq, STATE == "") |>
select(STATE, LOCATION_NAME)
## # A tibble: 9 × 2
## STATE LOCATION_NAME
## <chr> <chr>
## 1 "" MONTANA: CLARKSTON VALLEY
## 2 "" ILLINOIS: WEST SALEM
## 3 "" NEVADA: FALLON
## 4 "" CALIFORNIA: OCOTILLO
## 5 "" COLORADO: PAONIA
## 6 "" ALASKA: ALEUTIAN ISLANDS: FOX ISLANDS
## 7 "" OKLAHOMA: SPARKS
## 8 "" OKLAHOMA: SPARKS, PRAGUE
## 9 "" TEXAS: WESTOne way to fix this:
badidx <- which(eq$STATE == "")
badstate <- sub(":.*", "",
eq$LOCATION_NAME[badidx])
eq$STATE[badidx] <-
state.abb[match(tolower(badstate),
tolower(state.name))]Add full state names as a factor:
states <- data.frame(STATE = state.abb,
STATE_NAME = state.name)
eq <- left_join(eq, states, "STATE")
eq <- mutate(eq,
STATE_NAME = factor(STATE_NAME,
state.name))Look at total number of earthquakes by state:
tbl <- count(eq, STATE_NAME)
head(tbl, 6)
## # A tibble: 6 × 2
## STATE_NAME n
## <fct> <int>
## 1 Alabama 1
## 2 Alaska 78
## 3 Arkansas 2
## 4 California 100
## 5 Colorado 2
## 6 Connecticut 1library(forcats)
pb <- ggplot(tbl, aes(y = fct_rev(STATE_NAME), x = n)) +
geom_col() +
thm +
theme(axis.title.y = element_blank())
ps <- ggplot(tbl, aes(y = fct_rev(STATE_NAME), x = n)) +
geom_point() +
geom_segment(aes(xend = 0, yend = fct_rev(STATE_NAME))) +
thm +
theme(axis.title.y = element_blank())
pb | ps
Many states, including Iowa, are missing.
Their counts should be zero.
This often impacts visualizations.
complete() can be used to fill in the zero values.
tbl <-
count(eq, STATE_NAME) |>
complete(STATE_NAME, fill = list(n = 0))
tbl
## # A tibble: 50 × 2
## STATE_NAME n
## <fct> <int>
## 1 Alabama 1
## 2 Alaska 78
## 3 Arizona 0
## 4 Arkansas 2
## 5 California 100
## 6 Colorado 2
## 7 Connecticut 1
## 8 Delaware 0
## 9 Florida 0
## 10 Georgia 0
## # ℹ 40 more rowsthm_ytxt_sm <- theme(axis.text.y = element_text(size = 10))
(pb %+% tbl + thm_ytxt_sm) | (ps %+% tbl + thm_ytxt_sm)
Interactive Tutorial
An interactive learnr tutorial for these notes is available.
You can run the tutorial with
STAT4580::runTutorial("dplyr")You can install the current version of the STAT4580 package with
remotes::install_gitlab("luke-tierney/STAT4580")You may need to install the remotes package from CRAN first.
Exercises
To bring in
dplyrand thempgdata, start by evaluatinglibrary(dplyr) library(ggplot2)The
selectfunction allows variables to be specified in a variety of ways. Which of the following does not produce a data frame with only the variablesmanufacturer,model,cty,hwy?select(mpg, 1:2, 7:8)select(mpg, starts_with("m"), ends_with("y"))select(mpg, 1:2, cty : hwy)select(mpg, -(displ : drv), -(fl : class))
Consider the code
library(dplyr) library(ggplot2) filter(mpg, ---) |> nrow()Which of the replacements for
---computes the number of Ford vehicles with more than 6 cylinders in thempgtable?model == "ford", cyl <= 6manufacturer == "ford" | cyl > 6manufacturer == "ford", cyl > 6manufacturer == "ford", cylinders > 6
In the 2013 NYC flights data provided by the
nycflights13package how many flights were there to Des Moines (FAA code DSM) from NYC in the first three months of 2013?- 13
- 98
- 64
- 77
To bring in
dplyrand thempgdata, start by evaluatinglibrary(dplyr) library(ggplot2)Which of the following sorts the rows for
mpgby increasingcylvalue and, within eachcylvalue sorts the rows from largest to smallesthwyvalue.arrange(mpg, desc(hwy), cyl)arrange(mpg, cyl, desc(hwy))arrange(mpg, desc(cyl), hwy)arrange(mpg, cyl, hwy)
To bring in
dplyrand theflightsdata, start by evaluatinglibrary(dplyr) library(nycflights13)The
dep_timevariable in theflightsdata set from thenycflights13package is convenient to look at (529 means 5:29 AM and 2212 means 10:12 PM), but hard to compute with. Which of the following adds variablesdep_houranddep_mincontaining hour and minute of the departure time?mutate(flights, dep_hour = dep_time %/% 60, dep_min = dep_time %% 60)mutate(flights, dep_hour = dep_time %/% 100, dep_min = dep_time %% 100)mutate(flights, dep_hour = dep_time / 100, dep_min = dep_time - dep_hour)mutate(flights, dep_hour = dep_time %/% 60, dep_min = dep_hour %% 60)
Using the
gapminderdata set, the following code computes population totals for each continent and each year:library(dplyr) library(gapminder) cpops <- group_by(gapminder, continent, year) |> summarize(pop = sum(pop)) |> ungroup()To produce a plot to compare population growth over the years for the continents it is useful to standardize the population data for each continent, for example by dividing the population values by the average population size for each continent. One way to do this is with a grouped
mutate. The first line of your result should behead(cpops_std, 1) ## # A tibble: 1 × 4 ## continent year pop stdpop ## <fct> <int> <dbl> <dbl> ## 1 Africa 1952 237640501 0.461library(ggplot2) ggplot(cpops_std, aes(x = year, y = stdpop, color = continent)) + geom_line()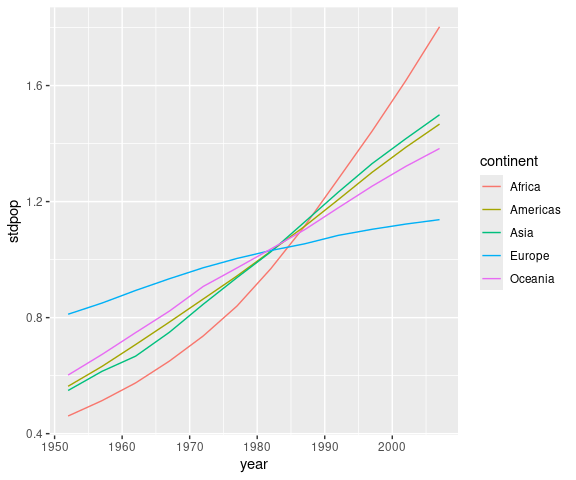
Which of the following produces the correct result:
cpops_std <- group_by(cpops, continent) |> mutate(stdpop = pop / mean(year)) |> ungroup()cpops_std <- group_by(cpops, year) |> mutate(stdpop = pop / mean(pop)) |> ungroup()cpops_std <- group_by(cpops, continent) |> mutate(stdpop = pop / mean(pop)) |> ungroup()cpops_std <- group_by(cpops, year) |> mutate(stdpop = pop / mean(year)) |> ungroup()
Another approach to the previous exercise first creates a table of population averages with
cpops_avg <- group_by(cpops, continent) |> summarize(avg_pop = mean(pop))Then use a left join to add
avg_popto thecpopstable, followed by an ordinary mutate step:left_join(---) |> mutate(stdpop = pop / avg_pop)Which is the correct replacement for
---?cpops_avg, cpops, "continent"cpops, cpops_avg, "continent"cpops, cpops_avg, "year"cpops_avg, cpops, "year"
