Snowfall in Iowa City
Adapted from a blog post by Sam Tyner
library(dplyr)
library(tidyr)
##library(rnoaa)
library(ggplot2)
library(ggrepel)
library(patchwork)
theme_set(theme_minimal() + theme(text = element_text(size = 16)))Getting the Data from NOAA
Climate data is available via a web service API from NOAA.
The rnoaa package provides an interface to this API. A tutorial is available from rOpenSci.
This code would be used to find the most suitable Iowa City station:
options("<key>") ## may not be needed
stat <- ghcnd_stations()
ic_stat <- filter(stat, grepl("IOWA CITY", name) & element == "SNOW")
select(ic_stat, name, id, ends_with("year"))The best station seems to be
ic_id <- "USC00134101"The current data for this station can then be obtained from NOAA:
ic_data_raw <- ghcnd(ic_id, refresh = TRUE)
## goes into a cache file as csv
## /home/luke/.cache/rnoaa/ghcnd/USC00134101.dly
write.csv(ic_data_raw, row.names = FALSE, file = "ic_noaa.csv")
## gzip and move to data directoryI have done these steps and stored the data locally; I will update the data as the course progresses.
Download the local data copy:
if (! file.exists("ic_noaa.csv.gz"))
download.file("http://www.stat.uiowa.edu/~luke/data/ic_noaa.csv.gz",
"ic_noaa.csv.gz")
## make sure data is up to date
ic_data_raw <- as_tibble(read.csv("ic_noaa.csv.gz", head = TRUE))Processing the Data
The raw data:
ic_data_raw
## # A tibble: 10,133 × 128
## id year month element VALUE1 MFLAG1 QFLAG1 SFLAG1 VALUE2 MFLAG2 QFLAG2
## <chr> <int> <int> <chr> <int> <chr> <chr> <chr> <int> <chr> <chr>
## 1 USC0013… 1893 1 TMAX -17 " " " " 6 -28 " " " "
## 2 USC0013… 1893 1 TMIN -67 " " " " 6 -161 " " " "
## 3 USC0013… 1893 1 PRCP 0 "P" " " 6 0 "P" " "
## 4 USC0013… 1893 1 SNOW 0 " " " " 6 0 " " " "
## 5 USC0013… 1893 2 TMAX 11 " " " " 6 -56 " " " "
## 6 USC0013… 1893 2 TMIN -233 " " " " 6 -206 " " " "
## 7 USC0013… 1893 2 PRCP 0 "P" " " 6 102 " " " "
## 8 USC0013… 1893 2 SNOW 0 " " " " 6 20 " " " "
## 9 USC0013… 1893 3 TMAX 56 " " " " 6 44 " " " "
## 10 USC0013… 1893 3 TMIN -78 " " " " 6 -67 " " " "
## # ℹ 10,123 more rows
## # ℹ 117 more variables: SFLAG2 <chr>, VALUE3 <int>, MFLAG3 <chr>, QFLAG3 <chr>,
## # SFLAG3 <chr>, VALUE4 <int>, MFLAG4 <chr>, QFLAG4 <chr>, SFLAG4 <chr>,
## # VALUE5 <int>, MFLAG5 <chr>, QFLAG5 <chr>, SFLAG5 <chr>, VALUE6 <int>,
## # MFLAG6 <chr>, QFLAG6 <chr>, SFLAG6 <chr>, VALUE7 <int>, MFLAG7 <chr>,
## # QFLAG7 <chr>, SFLAG7 <chr>, VALUE8 <int>, MFLAG8 <chr>, QFLAG8 <chr>,
## # SFLAG8 <chr>, VALUE9 <int>, MFLAG9 <chr>, QFLAG9 <chr>, SFLAG9 <chr>, …The documentation for the data is available at
The available elements, or measurements, are:
unique(ic_data_raw$element)
## [1] "TMAX" "TMIN" "PRCP" "SNOW" "WT16" "WT01" "TOBS" "SNWD" "WT18" "WT03"
## [11] "WT04" "WT14" "EVAP" "WDMV" "WT05" "DAEV" "MDEV" "DAWM" "MDWM" "DAPR"
## [21] "MDPR" "MNPN" "MXPN" "WESD"The primary ones are:
PRCP = Precipitation (tenths of mm)
SNOW = Snowfall (mm)
SNWD = Snow depth (mm)
TMAX = Maximum temperature (tenths of degrees C)
TMIN = Minimum temperature (tenths of degrees C)The values for day \(k\) are stored in the variable VALUE\(k\). A selection:
ic_feb2019 <- filter(ic_data_raw, year == 2019, month == 2)
select(ic_feb2019, element, VALUE1, VALUE28, VALUE31)
## # A tibble: 6 × 4
## element VALUE1 VALUE28 VALUE31
## <chr> <int> <int> <int>
## 1 TMAX -167 -72 NA
## 2 TMIN -344 -139 NA
## 3 TOBS -194 -128 NA
## 4 PRCP 18 0 NA
## 5 SNOW 51 0 NA
## 6 SNWD 330 51 NAWe need to:
- Reshape the data.
- Deal with the bogus days.
Start by selecting the columns we need:
ic_data <- select(ic_data_raw, year, month, element, starts_with("VALUE"))
ic_data
## # A tibble: 10,133 × 34
## year month element VALUE1 VALUE2 VALUE3 VALUE4 VALUE5 VALUE6 VALUE7 VALUE8
## <int> <int> <chr> <int> <int> <int> <int> <int> <int> <int> <int>
## 1 1893 1 TMAX -17 -28 -150 -44 -17 -106 -56 -67
## 2 1893 1 TMIN -67 -161 -233 -156 -156 -206 -111 -211
## 3 1893 1 PRCP 0 0 0 64 0 38 0 0
## 4 1893 1 SNOW 0 0 0 64 0 38 0 0
## 5 1893 2 TMAX 11 -56 -106 -122 33 28 -161 -94
## 6 1893 2 TMIN -233 -206 -217 -267 -122 -211 -256 -278
## 7 1893 2 PRCP 0 102 0 0 0 102 0 0
## 8 1893 2 SNOW 0 20 0 0 0 102 0 0
## 9 1893 3 TMAX 56 44 17 -28 17 61 83 72
## 10 1893 3 TMIN -78 -67 -122 -156 -122 -106 -28 17
## # ℹ 10,123 more rows
## # ℹ 23 more variables: VALUE9 <int>, VALUE10 <int>, VALUE11 <int>,
## # VALUE12 <int>, VALUE13 <int>, VALUE14 <int>, VALUE15 <int>, VALUE16 <int>,
## # VALUE17 <int>, VALUE18 <int>, VALUE19 <int>, VALUE20 <int>, VALUE21 <int>,
## # VALUE22 <int>, VALUE23 <int>, VALUE24 <int>, VALUE25 <int>, VALUE26 <int>,
## # VALUE27 <int>, VALUE28 <int>, VALUE29 <int>, VALUE30 <int>, VALUE31 <int>Reshape from (very) wide to (too) long:
ic_data <- pivot_longer(ic_data, VALUE1 : VALUE31,
names_to = "day", values_to = "value")
ic_data
## # A tibble: 314,123 × 5
## year month element day value
## <int> <int> <chr> <chr> <int>
## 1 1893 1 TMAX VALUE1 -17
## 2 1893 1 TMAX VALUE2 -28
## 3 1893 1 TMAX VALUE3 -150
## 4 1893 1 TMAX VALUE4 -44
## 5 1893 1 TMAX VALUE5 -17
## 6 1893 1 TMAX VALUE6 -106
## 7 1893 1 TMAX VALUE7 -56
## 8 1893 1 TMAX VALUE8 -67
## 9 1893 1 TMAX VALUE9 11
## 10 1893 1 TMAX VALUE10 -150
## # ℹ 314,113 more rowsExtract the day as a number:
ic_data <- mutate(ic_data, day = readr::parse_number(day))
ic_data
## # A tibble: 314,123 × 5
## year month element day value
## <int> <int> <chr> <dbl> <int>
## 1 1893 1 TMAX 1 -17
## 2 1893 1 TMAX 2 -28
## 3 1893 1 TMAX 3 -150
## 4 1893 1 TMAX 4 -44
## 5 1893 1 TMAX 5 -17
## 6 1893 1 TMAX 6 -106
## 7 1893 1 TMAX 7 -56
## 8 1893 1 TMAX 8 -67
## 9 1893 1 TMAX 9 11
## 10 1893 1 TMAX 10 -150
## # ℹ 314,113 more rowsReshape from too long to tidy with one row per day, keeping only the primary variables:
corevars <- c("TMAX", "TMIN", "PRCP", "SNOW", "SNWD")
ic_data <- filter(ic_data, element %in% corevars)
ic_data <- pivot_wider(ic_data, names_from = "element", values_from = "value")
ic_data
## # A tibble: 47,120 × 8
## year month day TMAX TMIN PRCP SNOW SNWD
## <int> <int> <dbl> <int> <int> <int> <int> <int>
## 1 1893 1 1 -17 -67 0 0 NA
## 2 1893 1 2 -28 -161 0 0 NA
## 3 1893 1 3 -150 -233 0 0 NA
## 4 1893 1 4 -44 -156 64 64 NA
## 5 1893 1 5 -17 -156 0 0 NA
## 6 1893 1 6 -106 -206 38 38 NA
## 7 1893 1 7 -56 -111 0 0 NA
## 8 1893 1 8 -67 -211 0 0 NA
## 9 1893 1 9 11 -144 0 0 NA
## 10 1893 1 10 -150 -250 0 0 NA
## # ℹ 47,110 more rowsAdd a date variable for plotting and to help get rid of bogus days:
ic_data <- mutate(ic_data, date = lubridate::make_date(year, month, day))
filter(ic_data, year == 2019, month == 2, day >= 27)
## # A tibble: 5 × 9
## year month day TMAX TMIN PRCP SNOW SNWD date
## <int> <int> <dbl> <int> <int> <int> <int> <int> <date>
## 1 2019 2 27 -44 -128 0 0 51 2019-02-27
## 2 2019 2 28 -72 -139 0 0 51 2019-02-28
## 3 2019 2 29 NA NA NA NA NA NA
## 4 2019 2 30 NA NA NA NA NA NA
## 5 2019 2 31 NA NA NA NA NA NAGet rid of the bogus days:
ic_data <- filter(ic_data, ! is.na(date))Make units more standard (American):
mm2in <- function(x) x / 25.4
C2F <- function(x) 32 + 1.8 * x
ic_data <- transmute(ic_data, year, month, day, date,
PRCP = mm2in(PRCP / 10),
SNOW = mm2in(SNOW),
SNWD = mm2in(SNWD),
TMIN = C2F(TMIN / 10),
TMAX = C2F(TMAX / 10))The Winters of 2018/2019 and 2020/2021
Daily Snowfall
Extract the data for days between November 1 and March 31:
w18 <- filter(ic_data, date >= "2018-11-01" & date <= "2019-03-31")
w20 <- filter(ic_data, date >= "2020-11-01" & date <= "2021-03-31")A first set of plots:
p1 <- ggplot(w18, aes(x = date)) + geom_col(aes(y = SNOW))
p2 <- ggplot(w18, aes(x = date)) + geom_line(aes(y = SNWD))
p1 + p2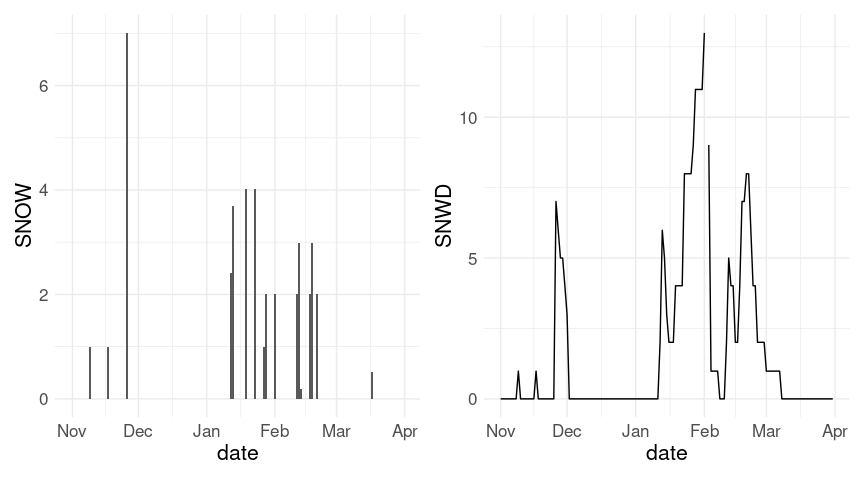
For daily snowfall it is probably reasonable to replace missing values by zeros.
w18 <- mutate(w18, SNOW = ifelse(is.na(SNOW), 0, SNOW))
w20 <- mutate(w20, SNOW = ifelse(is.na(SNOW), 0, SNOW))For snow depth it may make sense to fill forward, replacing misusing values by the previous non-missing value. Using the fill function from tidyr is one way to do this.
w18 <- fill(w18, SNWD)
w20 <- fill(w20, SNWD)The plot of snow depth looks a bit better with this change; the axis range for the plot of daily snowfall is also affected. Also, the warnings are eliminated.
p1 <- ggplot(w18, aes(x = date)) + geom_col(aes(y = SNOW))
p2 <- ggplot(w18, aes(x = date)) + geom_line(aes(y = SNWD))
(p1 + labs(title = "2018/19")) + p2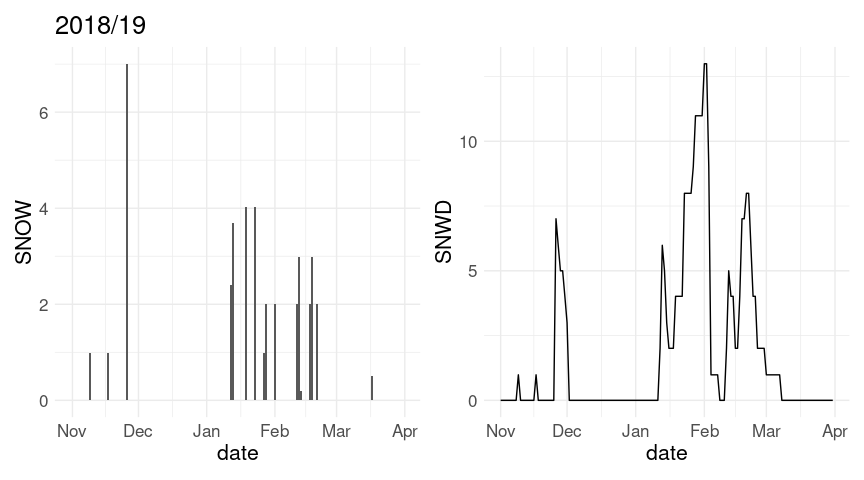
(p1 %+% w20 + labs(title = "2020/21")) + (p2 %+% w20)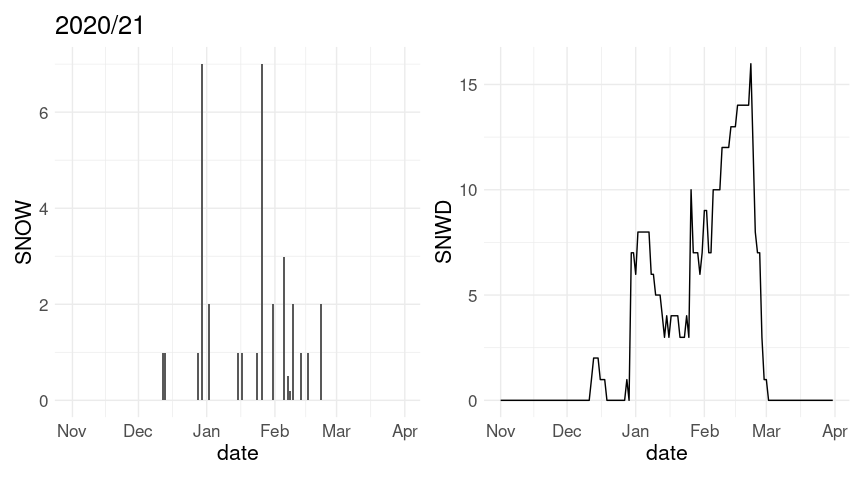
Often snowfall is visualized in terms of the cumulative snowfall for the season up to each date:
p <- ggplot(w18, aes(x = date)) + geom_line(aes(y = cumsum(SNOW)))
p + labs(title = "2018/19")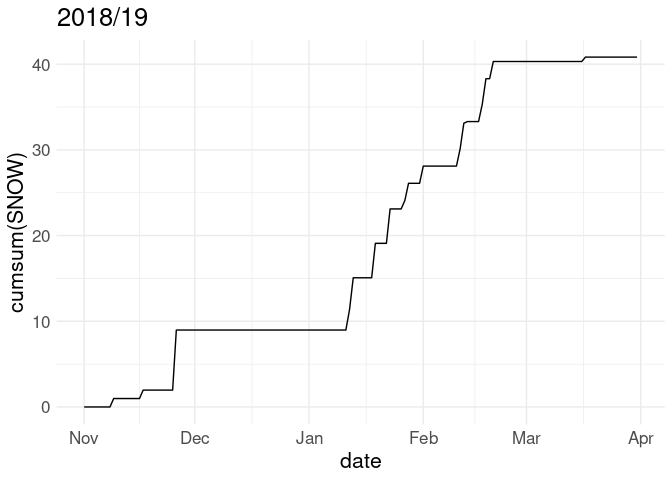
p %+% w20 + labs(title = "2020/21")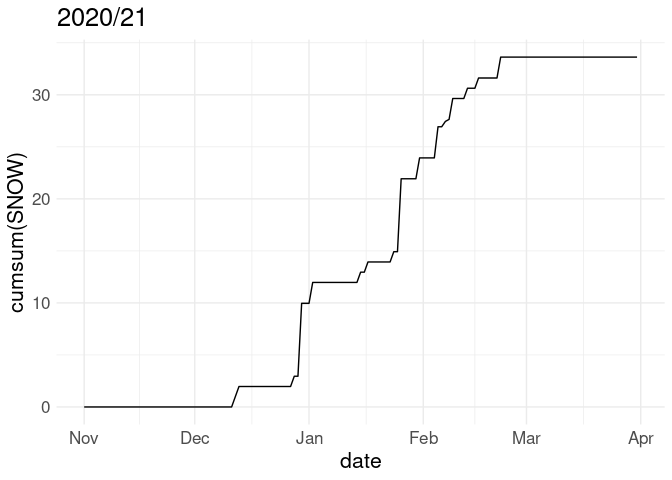
Comparing to Long Term Averages
Compute the average snowfall for each day of the year and merge the results for the days in w18 into w18 with a left_join:
ic_avg <- group_by(ic_data, month, day) |>
summarize(av_snow = mean(SNOW, na.rm = TRUE),
av_snwd = mean(SNWD, na.rm = TRUE)) |>
ungroup()
w18 <- left_join(w18, ic_avg, c("month", "day"))
w20 <- left_join(w20, ic_avg, c("month", "day"))Daily snowfall and snow cover for the 2018/19 winter along with averages over all years in the record:
p1 <- ggplot(w18, aes(x = date)) +
geom_col(aes(y = SNOW)) +
geom_line(aes(y = av_snow), color = "blue")
p2 <- ggplot(w18, aes(x = date)) +
geom_line(aes(y = SNWD)) +
geom_line(aes(y = av_snwd), color = "blue")
(p1 + labs(title = "2018/19")) + p2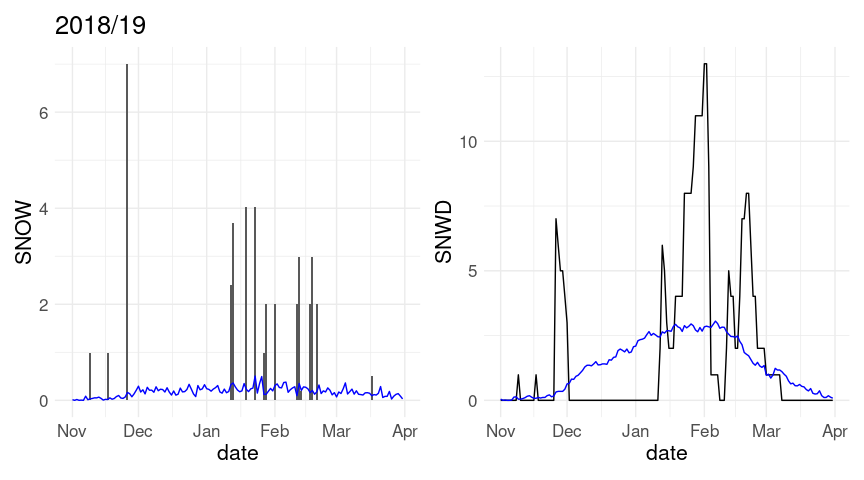
(p1 %+% w20 + labs(title = "2020/21")) + (p2 %+% w20)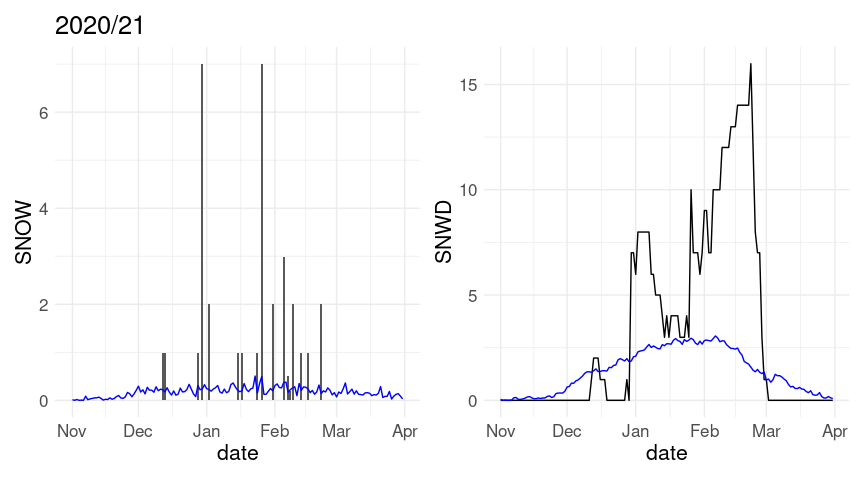
Cumulative snowfall for the 2018/19 winter along with average cumulative snowfall over all years in the record:
p <- ggplot(w18, aes(x = date)) +
geom_line(aes(y = cumsum(SNOW))) +
geom_line(aes(y = cumsum(av_snow)), color = "blue")
p + labs(title = "2018/19")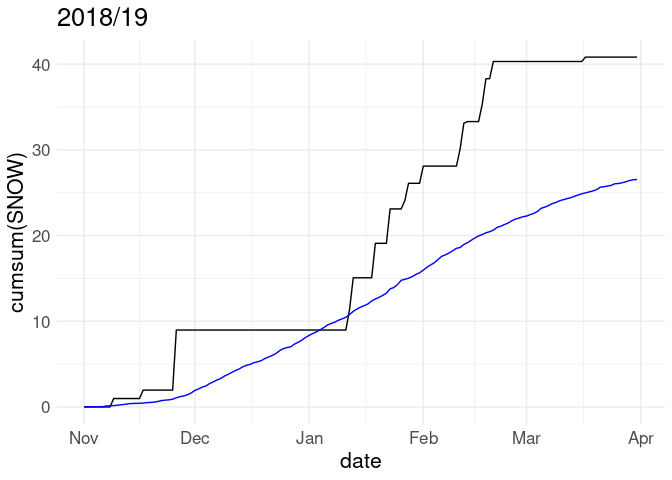
p %+% w20 + labs(title = "2020/21")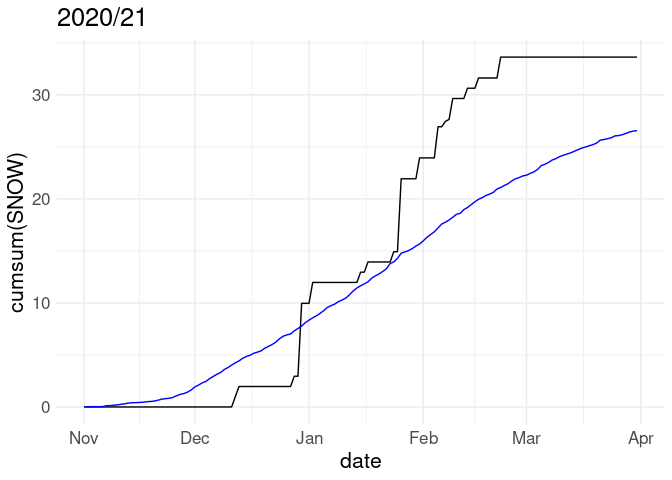
A Combined Graph
w18day <- transmute(w18, date, snow = SNOW,
base = "2018/19",
type = "Daily Snowfall")
nrmday <- transmute(w18, date, snow = av_snow,
base = "Normal",
type = "Daily Snowfall")
w18cum <- transmute(w18day, date, snow = cumsum(snow),
base = "2018/19",
type = "Cumulative Snowfall")
nrmcum <- transmute(nrmday, date, snow = cumsum(snow),
base = "Normal",
type = "Cumulative Snowfall")ggplot(w18, aes(x = date, y = snow, color = base)) +
geom_line(data = w18cum) +
geom_line(data = nrmcum) +
geom_segment(aes(x = date, xend = date, y = 0, yend = snow),
size = 1, data = w18day) +
geom_line(data = nrmday) +
facet_wrap(~ type, ncol = 1, scales = "free_y") +
xlab(element_blank()) + ylab("Snowfall (inches)") +
ggtitle("2018/19 Snowfall in Iowa City") +
scale_color_manual(values = c("2018/19" = "navy",
"Normal" = "forestgreen")) +
guides(color = guide_legend(override.aes = list(size = 1.5))) +
theme_bw() +
theme(legend.position = "bottom", legend.title = element_blank(),
plot.title = element_text(face = "bold", hjust = .5),
aspect.ratio = 0.4)
## Warning: Using `size` aesthetic for lines was deprecated in ggplot2 3.4.0.
## ℹ Please use `linewidth` instead.
## This warning is displayed once every 8 hours.
## Call `lifecycle::last_lifecycle_warnings()` to see where this warning was
## generated.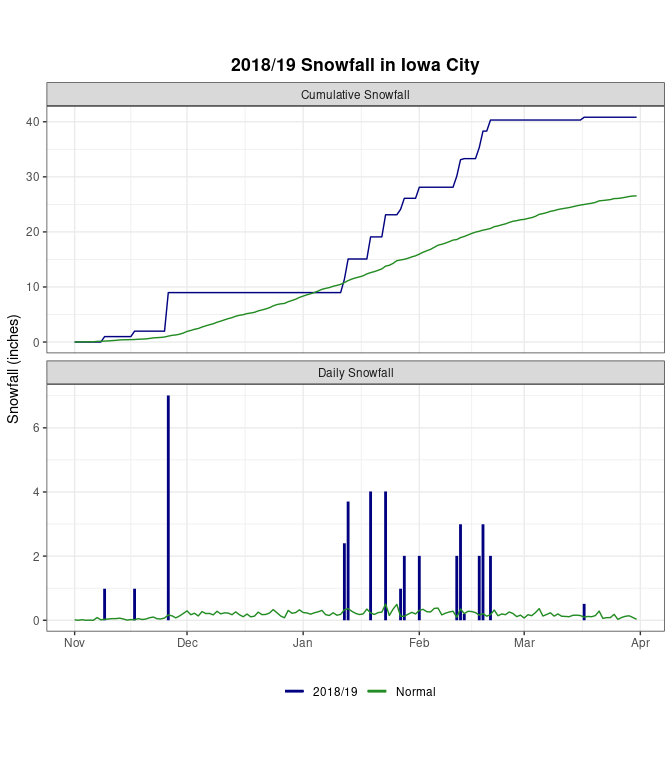
Total Snow Fall by Year and Month
It is helpful to define a variable that shifts the year to include a full winter, not the end of one and the start of the next.
What month should be start with? Here are the counts for months with recorded snowfall:
count(filter(ic_data, SNOW > 0), month)
## # A tibble: 9 × 2
## month n
## <int> <int>
## 1 1 559
## 2 2 456
## 3 3 300
## 4 4 73
## 5 5 1
## 6 7 1
## 7 10 20
## 8 11 136
## 9 12 477The July snowfall seems odd:
filter(ic_data, SNOW > 0, month == 7)
## # A tibble: 1 × 9
## year month day date PRCP SNOW SNWD TMIN TMAX
## <int> <int> <dbl> <date> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 1896 7 26 1896-07-26 0.110 2.91 NA 68 91.9This is probably a recording error.
For October, the years with recorded recorded snowfall are shown by
count(filter(ic_data, SNOW > 0, month == 10), year)
## # A tibble: 14 × 2
## year n
## <int> <int>
## 1 1898 1
## 2 1906 1
## 3 1913 2
## 4 1917 3
## 5 1923 1
## 6 1925 1
## 7 1929 1
## 8 1930 1
## 9 1967 1
## 10 1972 1
## 11 1980 2
## 12 1997 2
## 13 2019 2
## 14 2020 1It seems reasonable to start the winter year in October:
ic_data <- mutate(ic_data, wyear = ifelse(month >= 10, year, year - 1))The yearly total snowfall values:
tot <- group_by(ic_data, wyear) |>
summarize(tsnow = sum(SNOW, na.rm = TRUE))
tot18 <- filter(tot, wyear == 2018)
ggplot(tot, aes(x = wyear, y = tsnow)) +
geom_line() +
geom_point(data = tot18, color = "red") +
geom_text_repel(data = tot18, label = "2018/19", color = "red")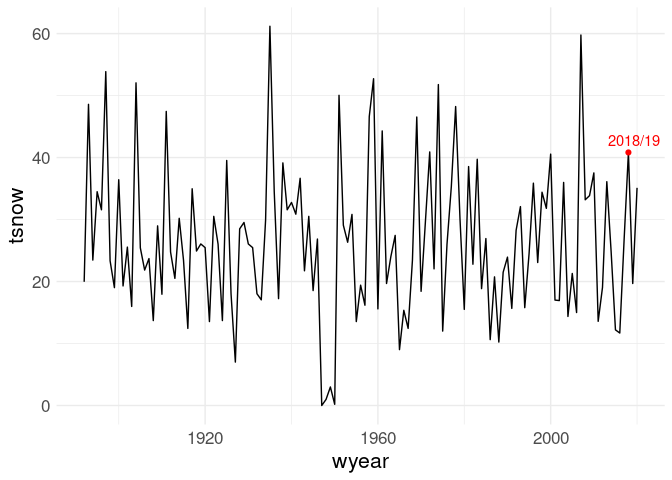
So 2018/19 is not a record, but still pretty high:
tot100 <- mutate(filter(tot, wyear >= 1918), rank = min_rank(desc(tsnow)))
filter(tot100, wyear == 2018)
## # A tibble: 1 × 3
## wyear tsnow rank
## <dbl> <dbl> <int>
## 1 2018 40.8 11What are the months with the highest average total snowfall?
mtot <- group_by(ic_data, year, wyear, month) |>
summarize(msnow = sum(SNOW, na.rm = TRUE)) |>
ungroup()
## `summarise()` has grouped output by 'year', 'wyear'. You can override using the
## `.groups` argument.
group_by(mtot, month) |> summarize(av_msnow = mean(msnow))
## # A tibble: 12 × 2
## month av_msnow
## <int> <dbl>
## 1 1 7.41
## 2 2 6.54
## 3 3 4.22
## 4 4 1.03
## 5 5 0.00153
## 6 6 0
## 7 7 0.0228
## 8 8 0
## 9 9 0
## 10 10 0.256
## 11 11 1.54
## 12 12 6.27For 2018/19:
filter(mtot, wyear == 2018)
## # A tibble: 12 × 4
## year wyear month msnow
## <int> <dbl> <int> <dbl>
## 1 2018 2018 10 0
## 2 2018 2018 11 8.98
## 3 2018 2018 12 0
## 4 2019 2018 1 17.1
## 5 2019 2018 2 14.2
## 6 2019 2018 3 0.512
## 7 2019 2018 4 0
## 8 2019 2018 5 0
## 9 2019 2018 6 0
## 10 2019 2018 7 0
## 11 2019 2018 8 0
## 12 2019 2018 9 0For 2020/21:
filter(mtot, wyear == 2020)
## # A tibble: 8 × 4
## year wyear month msnow
## <int> <dbl> <int> <dbl>
## 1 2020 2020 10 1.50
## 2 2020 2020 11 0
## 3 2020 2020 12 9.96
## 4 2021 2020 1 14.0
## 5 2021 2020 2 9.69
## 6 2021 2020 3 0
## 7 2021 2020 4 0
## 8 2021 2020 5 0A Data Processing Functions
If you want to look at data for another city you would need to go through the same steps starting with a new raw data frame.
Defining a function that encapsulates the steps makes this easy:
processData <- function(data_raw) {
## select just the columns we need:
data <- select(data_raw, year, month, element, starts_with("VALUE"))
## reshape from (very) wide to (too) long
data <- pivot_longer(data, VALUE1 : VALUE31,
names_to = "day", values_to = "value")
## extract the day as a number:
data <- mutate(data, day = readr::parse_number(day))
## reshape from too long to tidy with one row per day, keeping
## only the primary variables:
corevars <- c("TMAX", "TMIN", "PRCP", "SNOW", "SNWD")
data <- filter(data, element %in% corevars)
data <- pivot_wider(data, names_from = "element", values_from = "value")
## add a 'date' variable and get rid of bogus days
data <- mutate(data, date = lubridate::make_date(year, month, day))
data <- filter(data, ! is.na(date))
## make units more standard (American)
mm2in <- function(x) x / 25.4
C2F <- function(x) 32 + 1.8 * x
data <- transmute(data, year, month, day, date,
PRCP = mm2in(PRCP / 10),
SNOW = mm2in(SNOW),
SNWD = mm2in(SNWD),
TMIN = C2F(TMIN / 10),
TMAX = C2F(TMAX / 10))
## add winter year variable 'wyear' starting in October
data <- mutate(data, wyear = ifelse(month >= 10, year, year - 1))
data
}A quick sanity check:
stopifnot(identical(ic_data, processData(ic_data_raw)))Often it is a good idea to include a sanity check like this in a code chunk marked as include = FALSE.
Other Explorations
pivot_longer(w18, TMIN | TMAX, names_to = "which", values_to = "value") |>
ggplot(aes(x = date)) +
geom_line(aes(y = value, color = which))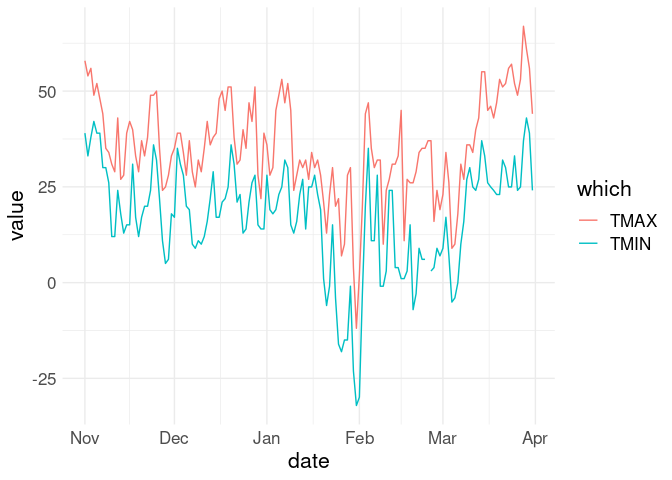
wd18 <- filter(ic_data, date >= "2018-10-01")
avs <- group_by(ic_data, month, day) |>
summarize(avsnow = mean(SNOW, na.rm = TRUE),
avsnwd = mean(SNWD, na.rm = TRUE),
avtmin = mean(TMIN, na.rm = TRUE),
avtmax = mean(TMAX, na.rm = TRUE)) |>
ungroup()
## `summarise()` has grouped output by 'month'. You can override using the
## `.groups` argument.
wd18 <- left_join(wd18, avs)
## Joining with `by = join_by(month, day)`
ggplot(filter(w18, ! is.na(SNWD))) +
geom_line(aes(x = date, y = SNWD)) +
geom_line(aes(x = date, y = av_snwd), col = "blue")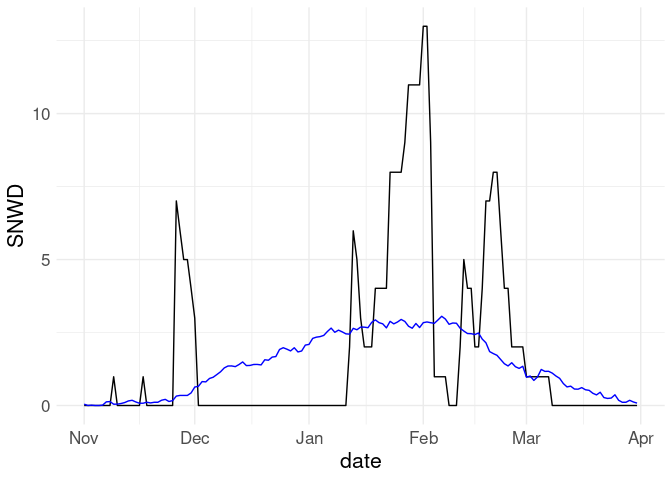
plot(SNOW ~ date, data = wd18, type = "h")
lines(wd18$date, wd18$avsnow, col = "red")
lines(smooth.spline(wd18$date, wd18$avsnow), col = "blue")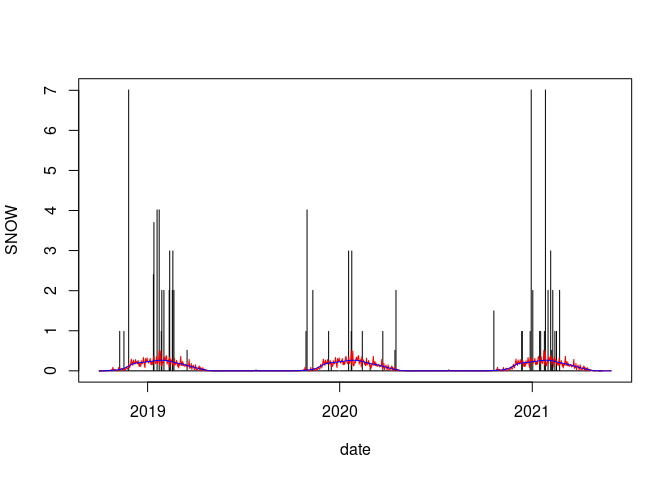
plot(SNWD ~ date, data = fill(wd18, SNWD), type = "l")
lines(wd18$date, wd18$avsnwd, col = "red")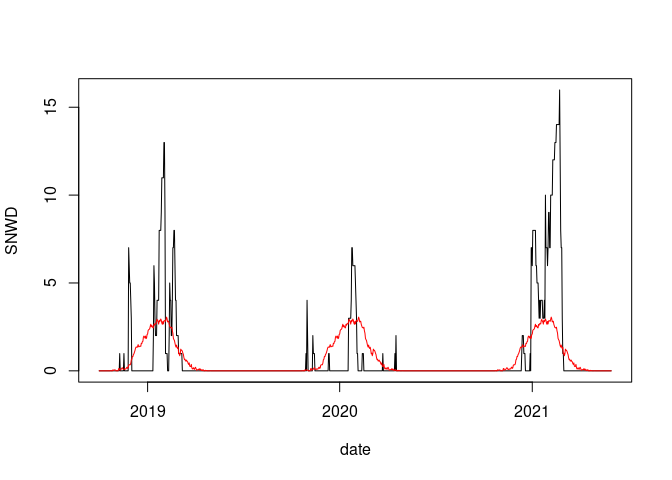
plot(cumsum(ifelse(is.na(SNOW), 0, SNOW)) ~ date, data = wd18, type = "l")
lines(wd18$date, cumsum(wd18$avsnow), col = "red")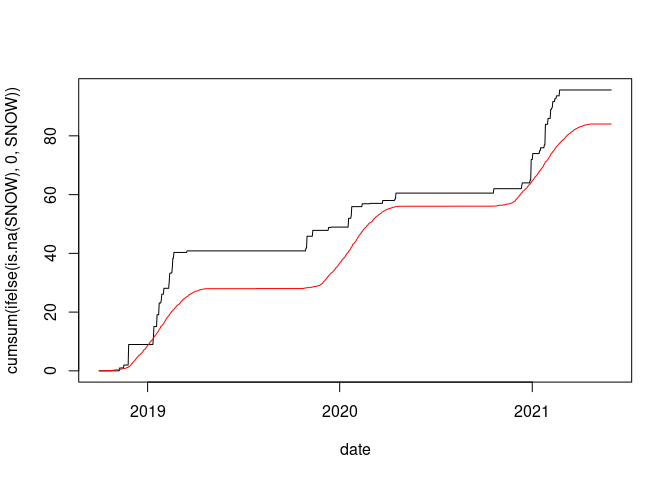
thm <- theme_bw() +
theme(panel.grid.major.x = element_blank(),
panel.grid.minor = element_blank())
ggplot(wd18) +
geom_col(aes(x = date, y = SNOW)) +
geom_line(aes(x = date, y = avsnow), col = "red") +
geom_smooth(aes(x = date, y = avsnow), se = FALSE) +
thm
## `geom_smooth()` using method = 'loess' and formula = 'y ~ x'
## Warning: Removed 25 rows containing missing values or values outside the scale range
## (`geom_col()`).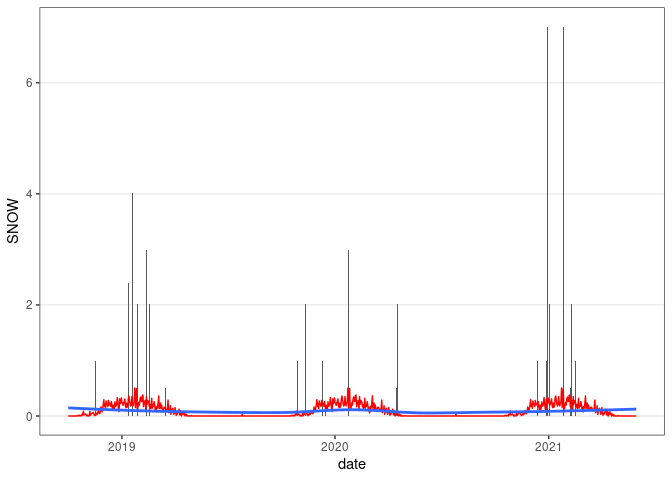
ggplot(fill(wd18, SNWD)) +
geom_line(aes(x = date, y = SNWD)) +
geom_line(aes(x = date, y = avsnwd), col = "red") +
geom_smooth(aes(x = date, y = avsnwd), se = FALSE) +
thm
## `geom_smooth()` using method = 'loess' and formula = 'y ~ x'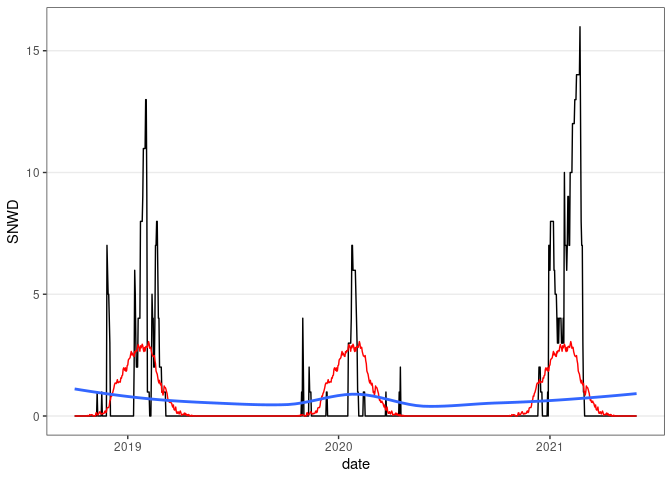
ggplot(wd18) +
geom_line(aes(x = date, y = cumsum(ifelse(is.na(SNOW), 0, SNOW)))) +
geom_line(aes(x = date, y = cumsum(wd18$avsnow)), col = "red") +
thm
## Warning: Use of `wd18$avsnow` is discouraged.
## ℹ Use `avsnow` instead.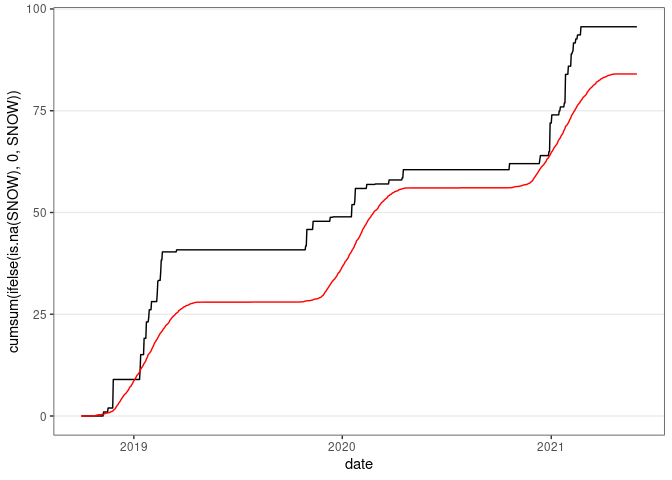
ggplot(filter(ic_data, year == 2019), aes(x = date)) +
geom_line(aes(y = TMAX, color = "TMAX")) +
geom_line(aes(y = TMIN, color = "TMIN")) +
thm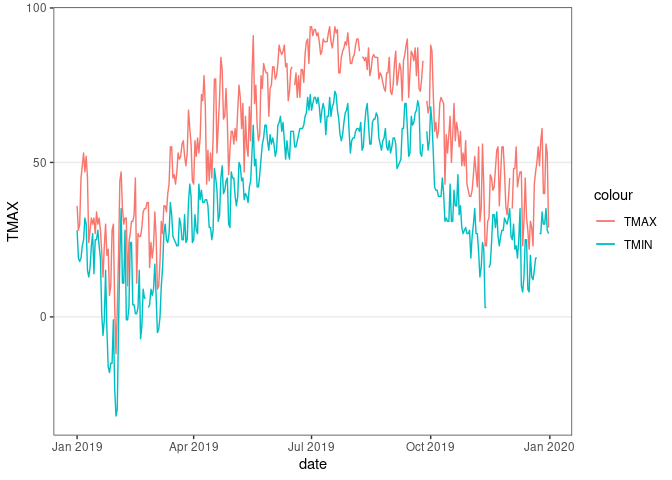
d13 <- filter(ic_data, wyear >= 2013, month <= 4 | month >= 10)
d13 <- mutate(d13, wday = as.numeric(date - lubridate::make_date(wyear, 10, 1)),
SNOW = ifelse(is.na(SNOW), 0, SNOW))
ggplot(d13) +
geom_col(aes(x = wday, y = SNOW), width = 3, position = "identity") +
facet_wrap(~ wyear) + thm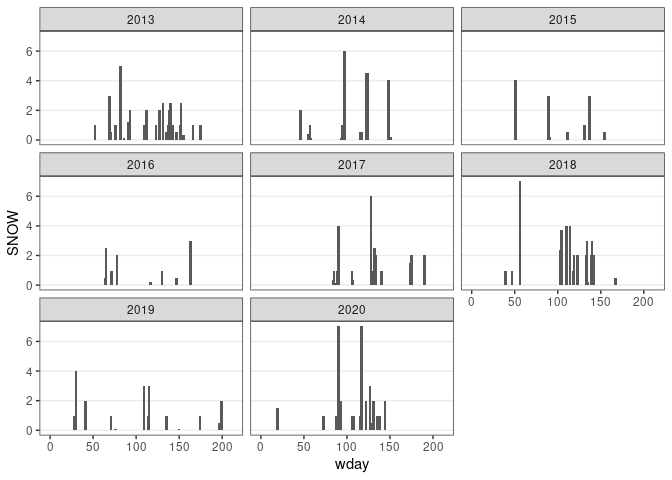
ggplot(d13) + geom_line(aes(x = wday, y = SNOW)) + facet_wrap(~ wyear) + thm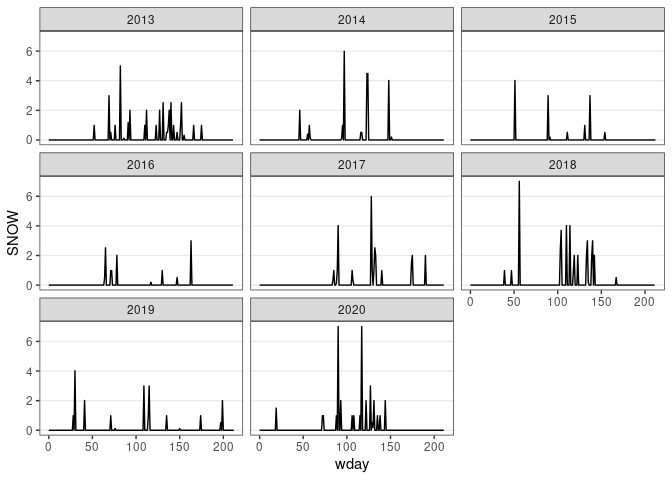
d13 <- group_by(d13, wyear) |>
mutate(snow_total = cumsum(SNOW)) |>
ungroup()
ggplot(d13) +
geom_line(aes(x = wday, y = snow_total, color = factor(wyear))) + thm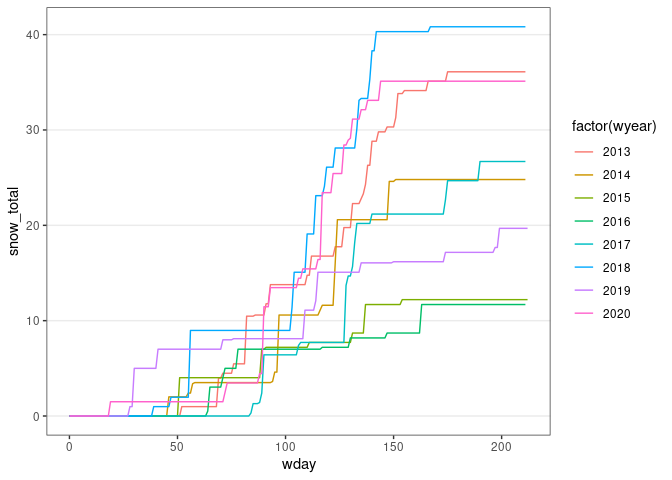
ggplot(d13) +
geom_line(aes(x = wday, y = snow_total)) +
facet_wrap(~ wyear) + thm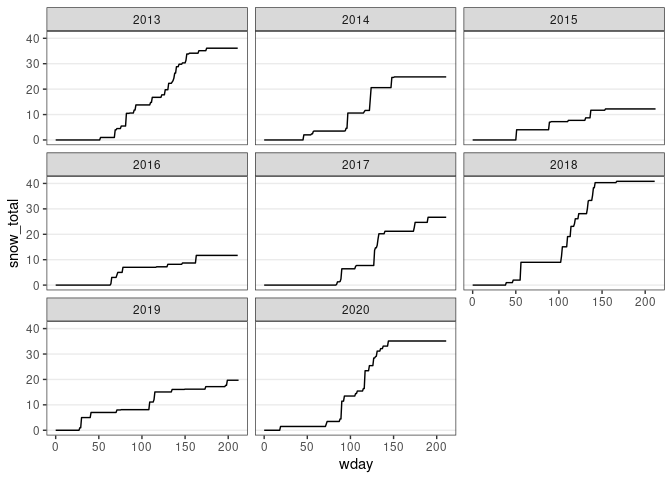
d13 <- fill(d13, SNWD)
ggplot(d13) + geom_line(aes(x = wday, y = SNWD)) + facet_wrap(~ wyear) + thm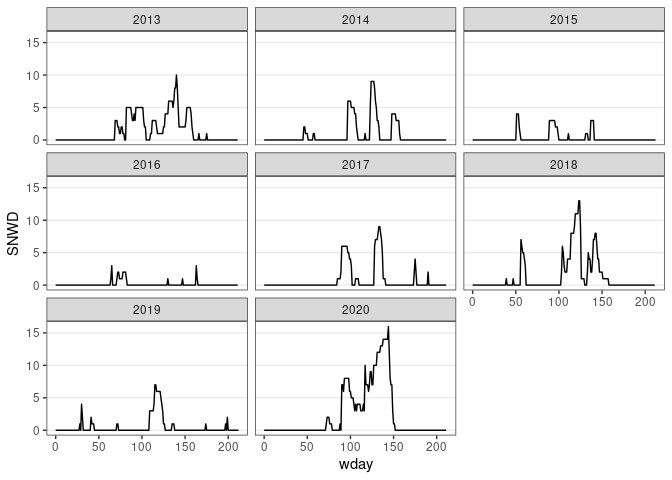
w <- filter(ic_data, wyear >= 2013)
w <- mutate(w, wday = date - lubridate::make_date(wyear, 10, 1))
w <- filter(w, wday <= 180)
w <- mutate(w, wday = lubridate::make_date(2018, 10, 1) + wday)
ggplot(w) +
geom_line(aes(x = wday, y = TMAX), color = "blue") +
geom_line(aes(x = wday, y = TMIN), color = "red") +
facet_wrap(~ wyear)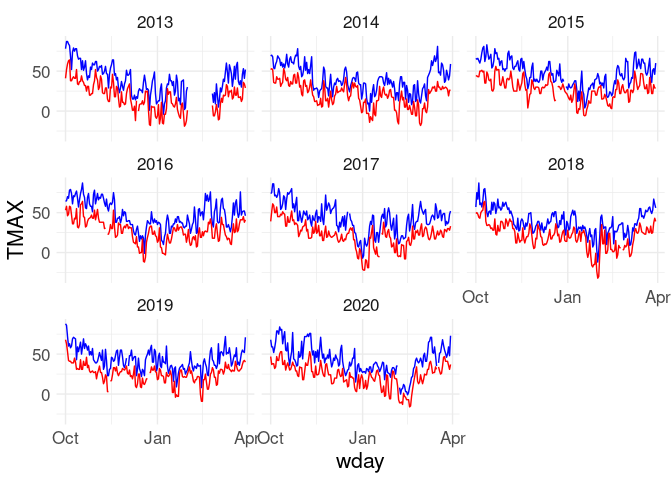
- Better x axis label for winter days
- set theme globally
- re-create original plot in some variants
- better facet labels
