Data Structures and Data Attribute Types
Data comes in many different forms.
Some of the most common data structures are
rectangular tables
networks and trees
geometries, regions
Other forms include
collections of text
video and audio recordings
…
We will work mostly with tables.
Many other forms can be reduced to tables.
Stevens (1945) classifies scales of measurement for attributes, or variables, as
nominal (e.g. hair color)
ordinal (e.g. dislike, neutral, like)
interval (e.g. temperature) — compare by difference
ratio (e.g. counts) — compare by ratio
These are sometimes grouped as
qualitative: nominal
quantitative: ordinal, interval, ratio
These can be viewed as semantic classifications
Computational considerations often classify variables as
categorical
integer, discrete
real, continuous
Another consideration is that some scales may be cyclic:
hours of the day
angles, longitude
These distinctions can be important in choosing visual representations.
Other typologies include one proposed by Mosteller and Tukey (1977) :
Names
Grades (ordered labels like beginner, intermediate, advanced)
Ranks (orders with 1 being the smallest or largest, 2 the next smallest or largest, and so on)
Counted fractions (bound by 0 and 1)
Counts (non-negative integers)
Amounts (non-negative real numbers)
Balances (any real number)
Data Types in R
R variables can be of different types.
The most common types are
numeric for real numbersintegercharacter for text data or nominal datafactor for nominal or ordinal data
Factors can be
unordered, for nominal data
ordered, for ordinal data
factors are more efficient and powerful for representing nominal or ordinal data than character data but can take a bit more getting used to.
Membership predicates and coercion functions are
is.numericas.numeric
is.integeras.integer
is.characteras.character
is.factoras.factor
is.orderedas.ordered
Conversion of factors with numeric-looking labels to numeric data should always go through as.character first.
Data Frames: Organizing Cases and Variables
Tabular data in R is usually stored as a data frame .
A data frame is a collection of variables , each with a value for every case or observacion .
The faithful data set is a data frame:
class(faithful)
## [1] "data.frame"
names(faithful)
## [1] "eruptions" "waiting"Most tools we work with in R use data organized in data frames.
Our plot() and lm() expressions from the introductory section can also we written as
plot(waiting ~ eruptions, data = faithful,
xlab = "Eruption time (min)",
ylab = "Waiting time to next eruption (min)")
fit <- lm(waiting ~ eruptions, data = faithful)
plot() only uses the data argument when the plot is specified as a formula , like
waiting ~ eruptions
Examining the Data in a Data Frame
head() provides an idea of what the raw data looks like:
head(faithful)
## eruptions waiting
## 1 3.600 79
## 2 1.800 54
## 3 3.333 74
## 4 2.283 62
## 5 4.533 85
## 6 2.883 55str() is also useful for an overview of an object’s structure:
str(faithful)
## 'data.frame': 272 obs. of 2 variables:
## $ eruptions: num 3.6 1.8 3.33 2.28 4.53 ...
## $ waiting : num 79 54 74 62 85 55 88 85 51 85 ...Another useful function available from the dplyr or tibble packages is glimpse():
library(dplyr)
glimpse(faithful)
## Rows: 272
## Columns: 2
## $ eruptions <dbl> 3.600, 1.800, 3.333, 2.283, 4.533, 2.883, 4.700, 3.600, 1.95…
## $ waiting <dbl> 79, 54, 74, 62, 85, 55, 88, 85, 51, 85, 54, 84, 78, 47, 83, …summary() shows basic statistical properties of the variables:
summary(faithful)
## eruptions waiting
## Min. :1.600 Min. :43.0
## 1st Qu.:2.163 1st Qu.:58.0
## Median :4.000 Median :76.0
## Mean :3.488 Mean :70.9
## 3rd Qu.:4.454 3rd Qu.:82.0
## Max. :5.100 Max. :96.0summary() with a character variable and a factor variable:
ffl <- mutate(faithful,
type = ifelse(eruptions < 3, "short", "long"),
ftype = factor(type))
summary(ffl)
## eruptions waiting type ftype
## Min. :1.600 Min. :43.0 Length:272 long :175
## 1st Qu.:2.163 1st Qu.:58.0 Class :character short: 97
## Median :4.000 Median :76.0 Mode :character
## Mean :3.488 Mean :70.9
## 3rd Qu.:4.454 3rd Qu.:82.0
## Max. :5.100 Max. :96.0
Variables in a Data Frame
A Data frame is a list, or vector, of variables:
length(faithful)
## [1] 2The dollar sign $ can be used to examine individual variables:
class(faithful$eruptions)
## [1] "numeric"
class(faithful$waiting)
## [1] "numeric"The variables can also be extracted by numerical or character index using the element extraction operation [[:
class(faithful[[1]])
## [1] "numeric"
class(faithful[["waiting"]])
## [1] "numeric"
Dimensions
The numbers of rows and columns can be obtained using nrow() and ncol():
ncol(faithful)
## [1] 2
nrow(faithful)
## [1] 272dim() returns a vector of the dimensions:
dim(faithful)
## [1] 272 2
Simple Visualizations
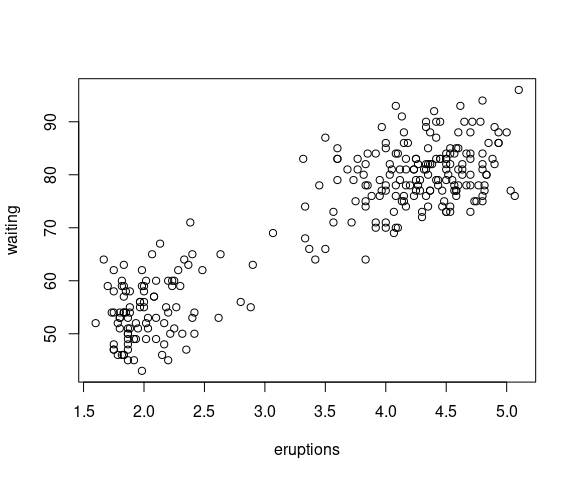
plot has a method for data frames that tries to provide a reasonable default visualization for numeric data frames:
plot(faithful)
Sample Data Sets
The datasets
Another package with a useful collection of data sets is dslabs
Many other data sets are contained in contributed packages as examples.
There are also many contributed packages designed specifically for making particular data sets available.
Tidy Data
The useful concept of a tidy data frame was introduced fairly recently and is described in a chapter in R for Data Science .
The idea is that
every observation should correspond to a single row;
every variable should correspond to a single column.
Tidy data is computationally convenient, and many of the tools we will use are designed around tidy data frames.
A large range of these tools can be accessed by loading the tidyverse package:
library(tidyverse)But for now I will load the needed packages individually.
The term tidy is a little unfortunate.
Data that is not tidy isn’t necessarily bad .
For human reading, and for some computations, data in a wider format can be better.
For other computations data in a longer, or narrower, format can be better.
Tibbles
Many tools in the tidyverse produce slightly enhanced data frames called tibbles :
library(tibble)
faithful_tbl <- as_tibble(faithful)
class(faithful_tbl)
## [1] "tbl_df" "tbl" "data.frame"Tibbles print differently from standard data frames:
faithful_tbl
## # A tibble: 272 × 2
## eruptions waiting
## <dbl> <dbl>
## 1 3.6 79
## 2 1.8 54
## 3 3.33 74
## 4 2.28 62
## 5 4.53 85
## 6 2.88 55
## 7 4.7 88
## 8 3.6 85
## 9 1.95 51
## 10 4.35 85
## # ℹ 262 more rowsFor the most part data frames and tibbles can be used interchangeably.
Tidying Data
Many data sets are in tidy form already.
If they are not, they can be put into tidy form.
The tools for this are part of data technologies .
The tasks involved are part of what is sometimes called data wrangling .
An Example: Global Average Surface Temperatures
Among many data sets available at https://data.giss.nasa.gov/gistemp/ is data on monthly global average surface temperatures over a number of years.
These data from 2017 were used for the widely cited Bloomberg hottest year visualization .
The current data are available in a formatted text file at
https://data.giss.nasa.gov/gistemp/tabledata_v4/GLB.Ts+dSST.txt
or as a CSV (comma-separated values)
https://data.giss.nasa.gov/gistemp/tabledata_v4/GLB.Ts+dSST.csv
The first few lines of the CSV file:
Land-Ocean: Global Means
Year,Jan,Feb,Mar,Apr,May,Jun,Jul,Aug,Sep,Oct,Nov,Dec,J-D,D-N,DJF,MAM,JJA,SON
1880,-.20,-.26,-.09,-.17,-.10,-.22,-.21,-.11,-.16,-.23,-.23,-.19,-.18,***,***,-.12,-.18,-.21
1881,-.20,-.16,.02,.03,.06,-.19,.00,-.05,-.16,-.22,-.19,-.08,-.10,-.11,-.18,.04,-.08,-.19
1882,.15,.13,.04,-.17,-.14,-.23,-.17,-.08,-.15,-.24,-.17,-.37,-.12,-.09,.07,-.09,-.16,-.19
1883,-.30,-.37,-.13,-.19,-.18,-.08,-.08,-.15,-.23,-.12,-.24,-.12,-.18,-.20,-.35,-.17,-.10,-.20The CSV file is a little easier (for a computer program) to read in, so we will work with that.
The numbers in the CSV file represent deviations in degrees Celcius from the average temperature for the base period 1951-1980.
The file available on January 16, 2025, is now available locally .
We can make sure it has been downloaded to our working directory with
if (! file.exists("GLB.Ts+dSST.csv"))
download.file("https://stat.uiowa.edu/~luke/data/GLB.Ts+dSST.csv",
"GLB.Ts+dSST.csv")Assuming this locally available file has been downloaded, we can read in the data and drop some columns we don’t need with
library(readr)
gast <- read_csv("GLB.Ts+dSST.csv", skip = 1)[1 : 13]
The function read_csv is from the readr package, which is part of the tidyverse.
An alternative is the base R function read.csv.
A look at the first few lines:
head(gast, 5)
## # A tibble: 5 × 13
## Year Jan Feb Mar Apr May Jun Jul Aug Sep Oct Nov Dec
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 1880 -0.2 -0.26 -0.09 -0.17 -0.1 -0.22 -0.21 -0.11 -0.16 -0.23 -0.23 -0.19
## 2 1881 -0.2 -0.16 0.02 0.03 0.06 -0.19 0 -0.05 -0.16 -0.22 -0.19 -0.08
## 3 1882 0.15 0.13 0.04 -0.17 -0.14 -0.23 -0.17 -0.08 -0.15 -0.24 -0.17 -0.37
## 4 1883 -0.3 -0.37 -0.13 -0.19 -0.18 -0.08 -0.08 -0.15 -0.23 -0.12 -0.24 -0.12
## 5 1884 -0.13 -0.09 -0.37 -0.41 -0.34 -0.35 -0.31 -0.28 -0.28 -0.25 -0.34 -0.31And the last few lines:
tail(gast, 5)
## # A tibble: 5 × 13
## Year Jan Feb Mar Apr May Jun Jul Aug Sep Oct Nov Dec
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 2020 1.18 1.24 1.18 1.12 1 0.91 0.89 0.86 0.96 0.87 1.09 0.79
## 2 2021 0.81 0.64 0.89 0.76 0.79 0.84 0.92 0.81 0.92 0.98 0.92 0.87
## 3 2022 0.91 0.89 1.04 0.83 0.84 0.92 0.94 0.95 0.89 0.97 0.73 0.8
## 4 2023 0.88 0.97 1.23 0.99 0.94 1.08 1.19 1.19 1.48 1.34 1.42 1.35
## 5 2024 1.24 1.44 1.39 1.31 1.16 1.24 1.2 1.3 1.23 1.33 1.29 1.26The print() method for tibbles abbreviates the output.
It is neater and provides some useful additional information on variable data types.
But it shows only the first few rows and may not explicitly show some columns.
If some columns are skipped, you can ask to see more by calling print() explicitly:
print(tail(gast), width = 100)
## # A tibble: 6 × 13
## Year Jan Feb Mar Apr May Jun Jul Aug Sep Oct Nov Dec
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 2019 0.93 0.95 1.17 1.01 0.86 0.9 0.94 0.95 0.93 0.99 0.98 1.11
## 2 2020 1.18 1.24 1.18 1.12 1 0.91 0.89 0.86 0.96 0.87 1.09 0.79
## 3 2021 0.81 0.64 0.89 0.76 0.79 0.84 0.92 0.81 0.92 0.98 0.92 0.87
## 4 2022 0.91 0.89 1.04 0.83 0.84 0.92 0.94 0.95 0.89 0.97 0.73 0.8
## 5 2023 0.88 0.97 1.23 0.99 0.94 1.08 1.19 1.19 1.48 1.34 1.42 1.35
## 6 2024 1.24 1.44 1.39 1.31 1.16 1.24 1.2 1.3 1.23 1.33 1.29 1.26The format with one column per month is compact and useful for viewing and data entry.
But it is not in tidy format since
the monthly temperature variable is spread over 12 columns;
the month variable is encoded in the column headings.
For obvious reasons this data format is often referred to as wide format .
The tidy, or long , format would have three variables: Year, Month, and Temp.
One way to put this data frame in tidy, or long, format uses pivot_longer() from the tidyr package:
library(tidyr)
lgast <- pivot_longer(gast,
-Year, ## specifies the columns to use -- all but Year
names_to = "Month",
values_to = "Temp")The first few rows of the result:
head(lgast)
## # A tibble: 6 × 3
## Year Month Temp
## <dbl> <chr> <dbl>
## 1 1880 Jan -0.2
## 2 1880 Feb -0.26
## 3 1880 Mar -0.09
## 4 1880 Apr -0.17
## 5 1880 May -0.1
## 6 1880 Jun -0.22During plotting it is likely that the Month variable will be converted to a factor .
By default, this will order levels alphabetically, which is not what we want:
levels(as.factor(lgast$Month))
## [1] "Apr" "Aug" "Dec" "Feb" "Jan" "Jul" "Jun" "Mar" "May" "Nov" "Oct" "Sep"We can guard against this by converting Month to a factor with the right levels now:
lgast <- mutate(lgast, Month = factor(Month, levels = month.abb))
levels(lgast$Month)
## [1] "Jan" "Feb" "Mar" "Apr" "May" "Jun" "Jul" "Aug" "Sep" "Oct" "Nov" "Dec"We can now use this tidy version of the data to create a static version of the Bloomberg hottest year visualization .
The basic framework is set up with
library(ggplot2)
p <- ggplot(lgast) +
ggtitle("Global Average Surface Temperatures") +
theme(plot.title = element_text(hjust = 0.5))
ggplot objects only produce output when they are printed.
To see the plot in p we need to print it, for example by using a line with only p.
p
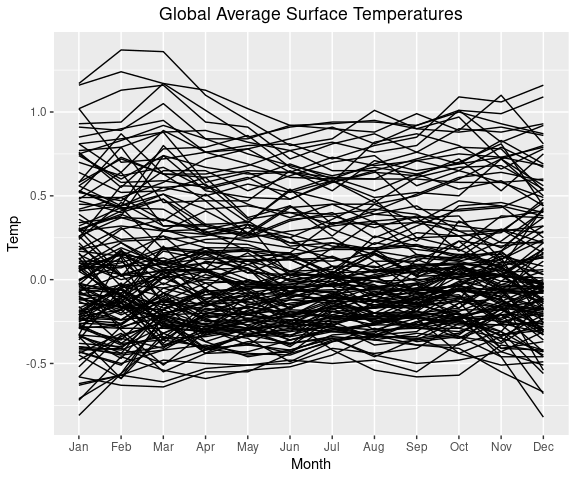
Then add a layer with lines for each year (specified by the group argument to geom_line).
p + geom_line(aes(x = Month,
y = Temp,
group = Year))
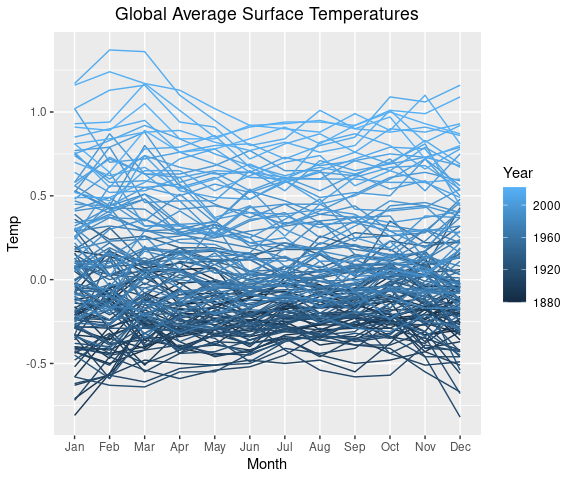
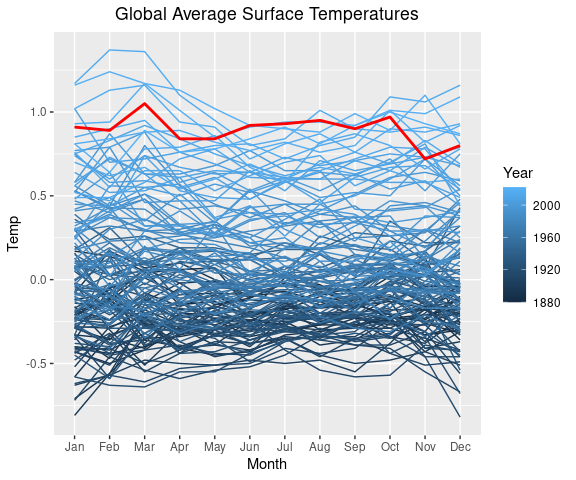
We can use color to distingish the years.
p1 <- p +
geom_line(aes(x = Month,
y = Temp,
color = Year,
group = Year),
na.rm = TRUE)
p1Saving the plot specification in the variable p1 will make it easier to experiment with color variations:
One way to highlight the past_year 2024:
lgast_last <- filter(lgast, Year == past_year)
p1 + geom_line(aes(x = Month,
y = Temp,
group = Year),
linewidth = 1,
color = "red",
data = lgast_last,
na.rm = TRUE)
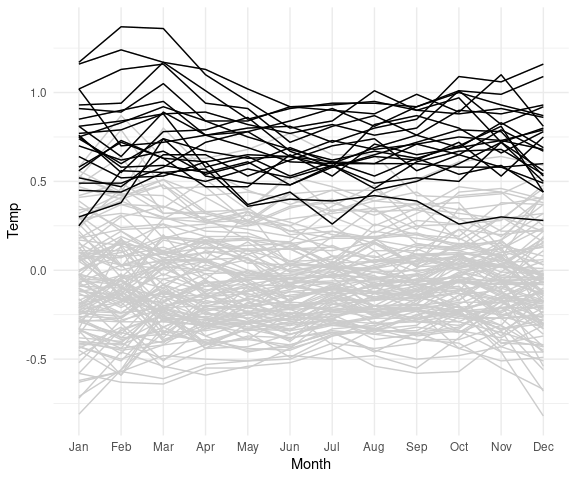
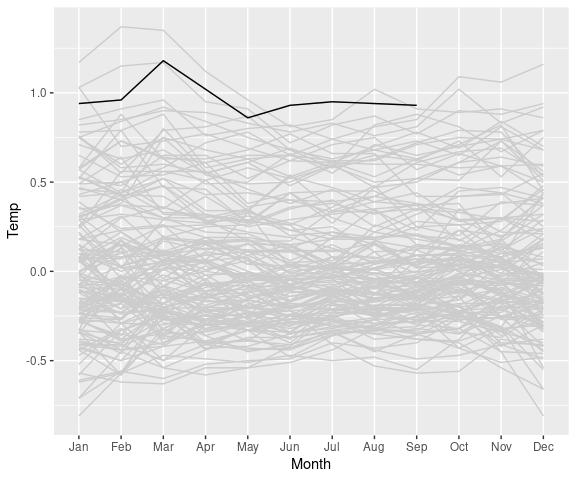
A useful way to show more recent data in the context of the full data is to show the full data in grey and the more recent years in black:
lgast2k <- filter(lgast, Year >= 2000)
ggplot(lgast, aes(x = Month,
y = Temp,
group = Year)) +
geom_line(color = "grey80") +
theme_minimal() +
geom_line(data = lgast2k)
If you want to update your plot later in the year then the current year’s entry may contain missing value indicators that you will have to deal with.
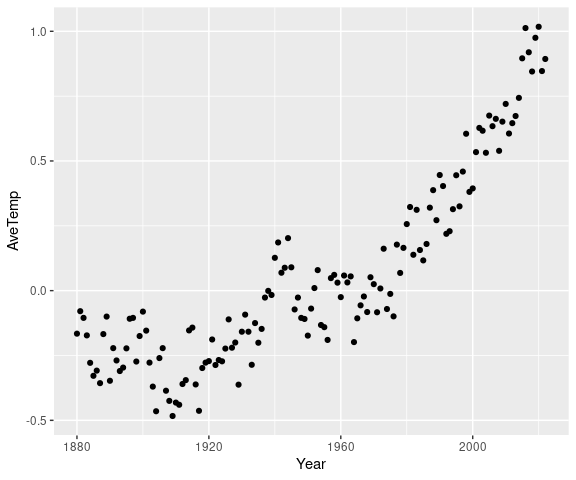
The New York Times on January 18, 2018, published another visualization of these data showing average yearly temperatures (via Google may work better ).
To recreate this plot we first need to compute yearly average temperatures.
This is easy to do with the summarize() and group_by() functions from dpyr:
library(dplyr)
atemp <- lgast |>
group_by(Year) |>
summarize(AveTemp = mean(Temp, na.rm = TRUE))
head(atemp)
## # A tibble: 6 × 2
## Year AveTemp
## <dbl> <dbl>
## 1 1880 -0.181
## 2 1881 -0.095
## 3 1882 -0.117
## 4 1883 -0.182
## 5 1884 -0.288
## 6 1885 -0.338Using na.rm = TRUE ensures that the mean is based on the available months if data for some months is missing.
A simple version of the plot is then produced by
ggplot(atemp) +
geom_point(aes(x = Year, y = AveTemp))
A variation showing record years:
library(ggrepel)
atemp_rec <- filter(atemp, cummax(AveTemp) == AveTemp)
ggplot(atemp, aes(x = Year, y = AveTemp)) +
geom_point() +
geom_point(data = atemp_rec, color = "red") +
geom_text_repel(aes(label = Year),
data = atemp_rec,
color = "blue")
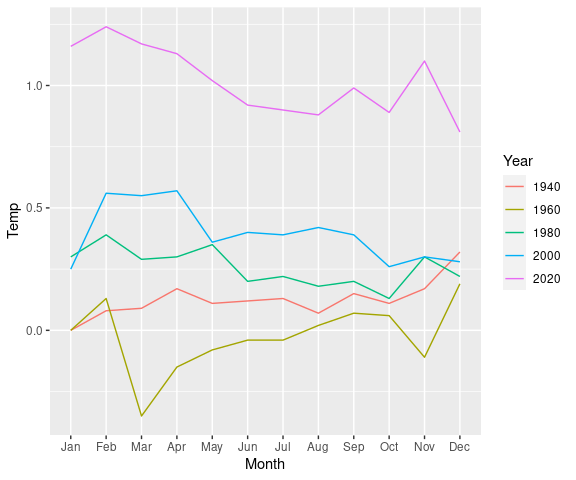
Another variation on the Bloomberg plot showing just a few years 20 years apart:
lg_by_20 <-
filter(lgast,
Year %in% seq(2020, by = -20, len = 5)) |>
mutate(Year = factor(Year))
ggplot(lg_by_20, aes(x = Month,
y = Temp,
group = Year,
color = Year)) +
geom_line()Converting Year to a factor results in a discrete color scale and legend.
Handling Missing Values
The data for 2019 available in early 2020 is also available locally .
Assuming this locally available file has been downloaded, we can read in the data and drop some columns we don’t need with
gast2019 <- read_csv("GLB.Ts+dSST-2019.csv", skip = 1)[1 : 13]The last three columns are read as character variables:
head(gast2019, 5)
## # A tibble: 5 × 13
## Year Jan Feb Mar Apr May Jun Jul Aug Sep Oct Nov Dec
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <chr> <chr> <chr>
## 1 1880 -0.17 -0.23 -0.08 -0.15 -0.08 -0.2 -0.17 -0.09 -0.13 -.22 -.20 -.16
## 2 1881 -0.18 -0.13 0.04 0.06 0.08 -0.17 0.02 -0.02 -0.14 -.20 -.17 -.05
## 3 1882 0.18 0.15 0.06 -0.15 -0.13 -0.21 -0.15 -0.06 -0.13 -.23 -.15 -.34
## 4 1883 -0.28 -0.36 -0.11 -0.17 -0.16 -0.07 -0.05 -0.12 -0.2 -.10 -.22 -.10
## 5 1884 -0.12 -0.07 -0.36 -0.39 -0.33 -0.34 -0.32 -0.27 -0.26 -.24 -.32 -.30The reason is that data for October through December were not available:
tail(gast2019, 2)
## # A tibble: 2 × 13
## Year Jan Feb Mar Apr May Jun Jul Aug Sep Oct Nov Dec
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <chr> <chr> <chr>
## 1 2018 0.82 0.85 0.9 0.89 0.83 0.78 0.83 0.76 0.81 1.02 .83 .92
## 2 2019 0.94 0.96 1.18 1.02 0.86 0.93 0.95 0.94 0.93 *** *** ***We want to convert these columns to numeric, with missing values represented as NA.
One option is to handle them individually:
gast2019$Oct <- as.numeric(gast2019$Oct)
## Warning: NAs introduced by coercion
tail(gast2019, 2)
## # A tibble: 2 × 13
## Year Jan Feb Mar Apr May Jun Jul Aug Sep Oct Nov Dec
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <chr> <chr>
## 1 2018 0.82 0.85 0.9 0.89 0.83 0.78 0.83 0.76 0.81 1.02 .83 .92
## 2 2019 0.94 0.96 1.18 1.02 0.86 0.93 0.95 0.94 0.93 NA *** ***Another option is to convert all character columns to numeric with
gast2019 <- mutate(gast2019, across(where(is.character), as.numeric))
## Warning: There were 2 warnings in `mutate()`.
## The first warning was:
## ℹ In argument: `across(where(is.character), as.numeric)`.
## Caused by warning:
## ! NAs introduced by coercion
## ℹ Run `dplyr::last_dplyr_warnings()` to see the 1 remaining warning.
tail(gast2019, 2)
## # A tibble: 2 × 13
## Year Jan Feb Mar Apr May Jun Jul Aug Sep Oct Nov Dec
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 2018 0.82 0.85 0.9 0.89 0.83 0.78 0.83 0.76 0.81 1.02 0.83 0.92
## 2 2019 0.94 0.96 1.18 1.02 0.86 0.93 0.95 0.94 0.93 NA NA NAThe warnings are benign and can be suppressed with the warning = FALSE chunk option.
Since we know the missing value pattern *** we can also avoid the need to fix the data after the fact by specifying this at read time:
gast2019 <- read_csv("GLB.Ts+dSST-2019.csv", na = "***", skip = 1)[1 : 13]
tail(gast2019, 2)
## # A tibble: 2 × 13
## Year Jan Feb Mar Apr May Jun Jul Aug Sep Oct Nov Dec
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 2018 0.82 0.85 0.9 0.89 0.83 0.78 0.83 0.76 0.81 1.02 0.83 0.92
## 2 2019 0.94 0.96 1.18 1.02 0.86 0.93 0.95 0.94 0.93 NA NA NAA plot highlighting the year 2019 shows only the months with available data:
lgast2019 <- gast2019 |>
pivot_longer(-Year,
names_to = "Month",
values_to = "Temp") |>
mutate(Month = factor(Month, levels = month.abb))
ggplot(lgast2019, aes(x = Month,
y = Temp,
group = Year)) +
geom_line(color = "grey80",
na.rm = TRUE) +
geom_line(data = filter(lgast2019, Year == 2019),
na.rm = TRUE)Adding na.rm = TRUE in the geom_line calls suppresses warnings; the plot would be the same without these.
Outline of the tools used:
Data processing:
Reading: read_csv, read.csv;
Reshaping: pivot_longer;
Cleaning: is.character, as.numeric, mutate, across, factor;
Summarizing: group_by, summarize.
Visualization geometries:
geom_line for a line plot;geom_point for a scatter plot.
Interactive Tutorial
An interactive learnravailable .
You can run the tutorial with
STAT4580::runTutorial("datafrm")
Exercises
Which of the Stevens classifications (nominal, ordinal, interval, ratio) best characterizes these variables:
Daily maximal temperatures in Iowa City.
Population counts for Iowa counties.
Education level of job applicants using the Bureau of Labor Statistics classification .
Major of UI students.
Which of these data sets are in tidy form?
The builtin data set co2
The builtin data set BOD
The who data set in package tidyr (tidyr::who)
The mpg data set in package ggplot2 (ggplot2::mpg)
The next exercises use the data in the variable gapminder in the package gapminder. You can make it available with
data(gapminder, package = "gapminder")
Use the function str to examine the value of the gapminder variable. How many cases are there in the data set? How many of the variables are factors?
Use the functions class and names to find the class and variable names in the gapminder data.
Use summary to compute summary information for the variables.
Fill in the values for --- needed to produce plots of life expectancy against year for the countries in continent Oceania.
library(dplyr)
library(ggplot2)
data(gapminder, package = "gapminder")
ggplot(filter(gapminder, continent == "Oceania"),
aes(x = ---, y = ---, color = country)) +
geom_line()
LS0tCnRpdGxlOiAiRGF0YSBhbmQgRGF0YSBGcmFtZXMiCm91dHB1dDoKICBodG1sX2RvY3VtZW50OgogICAgdG9jOiB5ZXMKICAgIGNvZGVfZG93bmxvYWQ6IHRydWUKLS0tCgo8bGluayByZWw9InN0eWxlc2hlZXQiIGhyZWY9InN0YXQ0NTgwLmNzcyIgdHlwZT0idGV4dC9jc3MiIC8+CgpgYGB7ciBzZXR1cCwgaW5jbHVkZT1GQUxTRX0Kc291cmNlKGhlcmU6OmhlcmUoInNldHVwLlIiKSkKa25pdHI6Om9wdHNfY2h1bmskc2V0KGNvbGxhcHNlID0gVFJVRSwgbWVzc2FnZSA9IEZBTFNFLAogICAgICAgICAgICAgICAgICAgICAgZmlnLmhlaWdodCA9IDUsIGZpZy53aWR0aCA9IDYsIGZpZy5hbGlnbiA9ICJjZW50ZXIiKQpvcHRpb25zKHdpZHRoID0gODApCmBgYAoKCiMjIERhdGEgU3RydWN0dXJlcyBhbmQgRGF0YSBBdHRyaWJ1dGUgVHlwZXMKCkRhdGEgY29tZXMgaW4gbWFueSBkaWZmZXJlbnQgZm9ybXMuCgpTb21lIG9mIHRoZSBtb3N0IGNvbW1vbiBkYXRhIHN0cnVjdHVyZXMgYXJlCgoqIHJlY3Rhbmd1bGFyIHRhYmxlcwoqIG5ldHdvcmtzIGFuZCB0cmVlcwoqIGdlb21ldHJpZXMsIHJlZ2lvbnMKCk90aGVyIGZvcm1zIGluY2x1ZGUKCiogY29sbGVjdGlvbnMgb2YgdGV4dAoqIHZpZGVvIGFuZCBhdWRpbyByZWNvcmRpbmdzCiogLi4uCgpXZSB3aWxsIHdvcmsgbW9zdGx5IHdpdGggdGFibGVzLgoKTWFueSBvdGhlciBmb3JtcyBjYW4gYmUgcmVkdWNlZCB0byB0YWJsZXMuCgpbU3RldmVuc10oaHR0cHM6Ly9lbi53aWtpcGVkaWEub3JnL3dpa2kvU3RhbmxleV9TbWl0aF9TdGV2ZW5zKSAoMTk0NSkKIGNsYXNzaWZpZXMgW3NjYWxlcyBvZgogbWVhc3VyZW1lbnRdKGh0dHBzOi8vZW4ud2lraXBlZGlhLm9yZy93aWtpL0xldmVsX29mX21lYXN1cmVtZW50KSBmb3IKIGF0dHJpYnV0ZXMsIG9yIHZhcmlhYmxlcywgYXMKPCEtLSBwZXJtYW5lbnQgbGluazogaHR0cHM6Ly9lbi53aWtpcGVkaWEub3JnL3cvaW5kZXgucGhwP3RpdGxlPUxldmVsX29mX21lYXN1cmVtZW50Jm9sZGlkPTEwNjA3NzI4NDcgLS0+IAoKKiBub21pbmFsIChlLmcuIGhhaXIgY29sb3IpCiogb3JkaW5hbCAoZS5nLiBkaXNsaWtlLCBuZXV0cmFsLCBsaWtlKQoqIGludGVydmFsIChlLmcuIHRlbXBlcmF0dXJlKSAtLS0gY29tcGFyZSBieSBkaWZmZXJlbmNlCiogcmF0aW8gKGUuZy4gY291bnRzKSAtLS0gY29tcGFyZSBieSByYXRpbwoKVGhlc2UgYXJlIHNvbWV0aW1lcyBncm91cGVkIGFzCgoqIHF1YWxpdGF0aXZlOiBub21pbmFsCiogcXVhbnRpdGF0aXZlOiBvcmRpbmFsLCBpbnRlcnZhbCwgcmF0aW8KClRoZXNlIGNhbiBiZSB2aWV3ZWQgYXMgX3NlbWFudGljIGNsYXNzaWZpY2F0aW9uc18KCl9Db21wdXRhdGlvbmFsIGNvbnNpZGVyYXRpb25zXyBvZnRlbiBjbGFzc2lmeSB2YXJpYWJsZXMgYXMKCiogY2F0ZWdvcmljYWwKKiBpbnRlZ2VyLCBkaXNjcmV0ZQoqIHJlYWwsIGNvbnRpbnVvdXMKCkFub3RoZXIgY29uc2lkZXJhdGlvbiBpcyB0aGF0IHNvbWUgc2NhbGVzIG1heSBiZSBjeWNsaWM6CgoqIGhvdXJzIG9mIHRoZSBkYXkKKiBhbmdsZXMsIGxvbmdpdHVkZQoKVGhlc2UgZGlzdGluY3Rpb25zIGNhbiBiZSBpbXBvcnRhbnQgaW4gY2hvb3NpbmcgdmlzdWFsIHJlcHJlc2VudGF0aW9ucy4KCk90aGVyIHR5cG9sb2dpZXMgaW5jbHVkZSBvbmUgcHJvcG9zZWQgYnkgW01vc3RlbGxlciBhbmQgVHVrZXkKKDE5NzcpXShodHRwczovL2VuLndpa2lwZWRpYS5vcmcvd2lraS9MZXZlbF9vZl9tZWFzdXJlbWVudCNNb3N0ZWxsZXJfYW5kX1R1a2V5J3NfdHlwb2xvZ3lfKDE5NzcpKToKCjEuIE5hbWVzCjIuIEdyYWRlcyAob3JkZXJlZCBsYWJlbHMgbGlrZSBiZWdpbm5lciwgaW50ZXJtZWRpYXRlLCBhZHZhbmNlZCkKMy4gUmFua3MgKG9yZGVycyB3aXRoIDEgYmVpbmcgdGhlIHNtYWxsZXN0IG9yIGxhcmdlc3QsIDIgdGhlIG5leHQKICAgc21hbGxlc3Qgb3IgbGFyZ2VzdCwgYW5kIHNvIG9uKQo0LiBDb3VudGVkIGZyYWN0aW9ucyAoYm91bmQgYnkgMCBhbmQgMSkKNS4gQ291bnRzIChub24tbmVnYXRpdmUgaW50ZWdlcnMpCjYuIEFtb3VudHMgKG5vbi1uZWdhdGl2ZSByZWFsIG51bWJlcnMpCjcuIEJhbGFuY2VzIChhbnkgcmVhbCBudW1iZXIpCgoKIyMgRGF0YSBUeXBlcyBpbiBSCgpSIHZhcmlhYmxlcyBjYW4gYmUgb2YgZGlmZmVyZW50IHR5cGVzLgoKVGhlIG1vc3QgY29tbW9uIHR5cGVzIGFyZQoKKiBgbnVtZXJpY2AgZm9yIHJlYWwgbnVtYmVycwoqIGBpbnRlZ2VyYAoqIGBjaGFyYWN0ZXJgIGZvciB0ZXh0IGRhdGEgb3Igbm9taW5hbCBkYXRhCiogYGZhY3RvcmAgZm9yIG5vbWluYWwgb3Igb3JkaW5hbCBkYXRhCgpGYWN0b3JzIGNhbiBiZQoKKiB1bm9yZGVyZWQsIGZvciBub21pbmFsIGRhdGEKKiBvcmRlcmVkLCBmb3Igb3JkaW5hbCBkYXRhCgo8ZGl2IGNsYXNzID0gImFsZXJ0Ij4KYGZhY3RvcnNgIGFyZSBtb3JlIGVmZmljaWVudCBhbmQgcG93ZXJmdWwgZm9yIHJlcHJlc2VudGluZyBub21pbmFsIG9yCm9yZGluYWwgZGF0YSB0aGFuIGBjaGFyYWN0ZXJgIGRhdGEgYnV0IGNhbiB0YWtlIGEgYml0IG1vcmUgZ2V0dGluZwp1c2VkIHRvLgo8L2Rpdj4KCk1lbWJlcnNoaXAgcHJlZGljYXRlcyBhbmQgY29lcmNpb24gZnVuY3Rpb25zIGFyZQoKfCBQcmVkaWNhdGUgICAgICAgfCBDb2Vyc2lvbnwKfDotLS0tLS0tLS0tLS0tLS0tfDotLS0tLS0tLS0tLS0tLS0tLXwKfCBgaXMubnVtZXJpY2AgICB8IGBhcy5udW1lcmljYCAgICB8CnwgYGlzLmludGVnZXJgICAgfCBgYXMuaW50ZWdlcmAgICAgfAp8IGBpcy5jaGFyYWN0ZXJgIHwgYGFzLmNoYXJhY3RlcmAgIHwKfCBgaXMuZmFjdG9yYCAgICB8IGBhcy5mYWN0b3JgICAgICB8CnwgYGlzLm9yZGVyZWRgICAgfCBgYXMub3JkZXJlZGAgICAgfAoKPGRpdiBjbGFzcyA9ICJhbGVydCI+CkNvbnZlcnNpb24gb2YgZmFjdG9ycyB3aXRoIG51bWVyaWMtbG9va2luZyBsYWJlbHMgdG8gbnVtZXJpYyBkYXRhCnNob3VsZCBhbHdheXMgZ28gdGhyb3VnaCBgYXMuY2hhcmFjdGVyYCBmaXJzdC4KPC9kaXY+CgoKIyMgRGF0YSBGcmFtZXM6IE9yZ2FuaXppbmcgQ2FzZXMgYW5kIFZhcmlhYmxlcwoKVGFidWxhciBkYXRhIGluIFIgaXMgdXN1YWxseSBzdG9yZWQgYXMgYSBfZGF0YSBmcmFtZV8uCgpBIGRhdGEgZnJhbWUgaXMgYSBjb2xsZWN0aW9uIG9mIF92YXJpYWJsZXNfLCBlYWNoIHdpdGggYSB2YWx1ZSBmb3IKZXZlcnkgX2Nhc2VfIG9yIF9vYnNlcnZhY2lvbl8uCgpUaGUgYGZhaXRoZnVsYCBkYXRhIHNldCBpcyBhIGRhdGEgZnJhbWU6CgpgYGB7cn0KY2xhc3MoZmFpdGhmdWwpCm5hbWVzKGZhaXRoZnVsKQpgYGAKCk1vc3QgdG9vbHMgd2Ugd29yayB3aXRoIGluIFIgdXNlIGRhdGEgb3JnYW5pemVkIGluIGRhdGEgZnJhbWVzLgoKT3VyIGBwbG90KClgIGFuZCBgbG0oKWAgZXhwcmVzc2lvbnMgZnJvbSB0aGUKW2ludHJvZHVjdG9yeSBzZWN0aW9uXShpbnRyby5odG1sKQpjYW4gYWxzbyB3ZSB3cml0dGVuIGFzCgpgYGB7ciwgZXZhbCA9IEZBTFNFfQpwbG90KHdhaXRpbmcgfiBlcnVwdGlvbnMsIGRhdGEgPSBmYWl0aGZ1bCwKICAgICB4bGFiID0gIkVydXB0aW9uIHRpbWUgKG1pbikiLAogICAgIHlsYWIgPSAiV2FpdGluZyB0aW1lIHRvIG5leHQgZXJ1cHRpb24gKG1pbikiKQpmaXQgPC0gbG0od2FpdGluZyB+IGVydXB0aW9ucywgZGF0YSA9IGZhaXRoZnVsKQpgYGAKCjxkaXYgY2xhc3MgPSAiYWxlcnQiPgpgcGxvdCgpYCBvbmx5IHVzZXMgdGhlIGBkYXRhYCBhcmd1bWVudCB3aGVuIHRoZSBwbG90IGlzIHNwZWNpZmllZCBhcyBhCl9mb3JtdWxhXywgbGlrZQoKYGBgcgp3YWl0aW5nIH4gZXJ1cHRpb25zCmBgYAo8L2Rpdj4KCgojIyBFeGFtaW5pbmcgdGhlIERhdGEgaW4gYSBEYXRhIEZyYW1lCgpgaGVhZCgpYCBwcm92aWRlcyBhbiBpZGVhIG9mIHdoYXQgdGhlIHJhdyBkYXRhIGxvb2tzIGxpa2U6CmBgYHtyfQpoZWFkKGZhaXRoZnVsKQpgYGAKCmBzdHIoKWAgaXMgYWxzbyB1c2VmdWwgZm9yIGFuIG92ZXJ2aWV3IG9mIGFuIG9iamVjdCdzIHN0cnVjdHVyZToKYGBge3J9CnN0cihmYWl0aGZ1bCkKYGBgCgpBbm90aGVyIHVzZWZ1bCBmdW5jdGlvbiBhdmFpbGFibGUgZnJvbSB0aGUgYGRwbHlyYCBvciBgdGliYmxlYApwYWNrYWdlcyBpcyBgZ2xpbXBzZSgpYDoKCmBgYHtyfQpsaWJyYXJ5KGRwbHlyKQpnbGltcHNlKGZhaXRoZnVsKQpgYGAKCmBzdW1tYXJ5KClgIHNob3dzIGJhc2ljIHN0YXRpc3RpY2FsIHByb3BlcnRpZXMgb2YgdGhlIHZhcmlhYmxlczoKCmBgYHtyfQpzdW1tYXJ5KGZhaXRoZnVsKQpgYGAKCmBzdW1tYXJ5KClgIHdpdGggYSBjaGFyYWN0ZXIgdmFyaWFibGUgYW5kIGEgZmFjdG9yIHZhcmlhYmxlOgoKYGBge3J9CmZmbCA8LSBtdXRhdGUoZmFpdGhmdWwsCiAgICAgICAgICAgICAgdHlwZSA9IGlmZWxzZShlcnVwdGlvbnMgPCAzLCAic2hvcnQiLCAibG9uZyIpLAogICAgICAgICAgICAgIGZ0eXBlID0gZmFjdG9yKHR5cGUpKQpzdW1tYXJ5KGZmbCkKYGBgCgoKIyMgVmFyaWFibGVzIGluIGEgRGF0YSBGcmFtZQoKQSBEYXRhIGZyYW1lIGlzIGEgbGlzdCwgb3IgdmVjdG9yLCBvZiB2YXJpYWJsZXM6CmBgYHtyfQpsZW5ndGgoZmFpdGhmdWwpCmBgYAoKVGhlIGRvbGxhciBzaWduIGAkYCBjYW4gYmUgdXNlZCB0byBleGFtaW5lIGluZGl2aWR1YWwgdmFyaWFibGVzOgpgYGB7cn0KY2xhc3MoZmFpdGhmdWwkZXJ1cHRpb25zKQpjbGFzcyhmYWl0aGZ1bCR3YWl0aW5nKQpgYGAKClRoZSB2YXJpYWJsZXMgY2FuIGFsc28gYmUgZXh0cmFjdGVkIGJ5IG51bWVyaWNhbCBvciBjaGFyYWN0ZXIgaW5kZXggdXNpbmcgdGhlCiAgZWxlbWVudCBleHRyYWN0aW9uIG9wZXJhdGlvbiBgW1tgOgpgYGB7cn0KY2xhc3MoZmFpdGhmdWxbWzFdXSkKY2xhc3MoZmFpdGhmdWxbWyJ3YWl0aW5nIl1dKQpgYGAKCgojIyBEaW1lbnNpb25zCgpUaGUgbnVtYmVycyBvZiByb3dzIGFuZCBjb2x1bW5zIGNhbiBiZSBvYnRhaW5lZCB1c2luZyBgbnJvdygpYCBhbmQgYG5jb2woKWA6CgpgYGB7cn0KbmNvbChmYWl0aGZ1bCkKbnJvdyhmYWl0aGZ1bCkKYGBgCgpgZGltKClgIHJldHVybnMgYSB2ZWN0b3Igb2YgdGhlIGRpbWVuc2lvbnM6CgpgYGB7cn0KZGltKGZhaXRoZnVsKQpgYGAKCgojIyBTaW1wbGUgVmlzdWFsaXphdGlvbnMKCmBwbG90YCBoYXMgYSBfbWV0aG9kXyBmb3IgZGF0YSBmcmFtZXMgdGhhdCB0cmllcyB0byBwcm92aWRlIGEKcmVhc29uYWJsZSBkZWZhdWx0IHZpc3VhbGl6YXRpb24gZm9yIG51bWVyaWMgZGF0YSBmcmFtZXM6CmBgYHtyfQpwbG90KGZhaXRoZnVsKQpgYGAKCgojIyBTYW1wbGUgRGF0YSBTZXRzCgpUaGUKW2BkYXRhc2V0c2BdKGh0dHA6Ly9zdGF0LmV0aHouY2gvUi1tYW51YWwvUi1kZXZlbC9saWJyYXJ5L2RhdGFzZXRzL2h0bWwvMDBJbmRleC5odG1sKQpwYWNrYWdlIGluIHRoZSBiYXNlIFIgZGlzdHJpYnV0aW9uIGNvbnRhaW5zIGEgbnVtYmVyIG9mIGRhdGEgc2V0cyB5b3UKY2FuIGV4cGxvcmUuCgpBbm90aGVyIHBhY2thZ2Ugd2l0aCBhIHVzZWZ1bCBjb2xsZWN0aW9uIG9mIGRhdGEgc2V0cyBpcyBbYGRzbGFic2BdKGh0dHBzOi8vY3Jhbi5yLXByb2plY3Qub3JnL3BhY2thZ2U9ZHNsYWJzKS4KCk1hbnkgb3RoZXIgZGF0YSBzZXRzIGFyZSBjb250YWluZWQgaW4gY29udHJpYnV0ZWQgcGFja2FnZXMgYXMgZXhhbXBsZXMuCgpUaGVyZSBhcmUgYWxzbyBtYW55IGNvbnRyaWJ1dGVkIHBhY2thZ2VzIGRlc2lnbmVkIHNwZWNpZmljYWxseSBmb3IKbWFraW5nIHBhcnRpY3VsYXIgZGF0YSBzZXRzIGF2YWlsYWJsZS4KCgojIyBUaWR5IERhdGEKClRoZSB1c2VmdWwgY29uY2VwdCBvZiBhIF90aWR5XyBkYXRhIGZyYW1lIHdhcyBpbnRyb2R1Y2VkIGZhaXJseQpbcmVjZW50bHldKGh0dHBzOi8vd3d3LmpzdGF0c29mdC5vcmcvYXJ0aWNsZS92aWV3L3YwNTlpMTApIGFuZCBpcwpkZXNjcmliZWQgaW4gYSBbY2hhcHRlciBpbiBfUiBmb3IgRGF0YQpTY2llbmNlX10oaHR0cHM6Ly9yNGRzLmhhZGxleS5uei9kYXRhLXRpZHkuaHRtbCkuCgpUaGUgaWRlYSBpcyB0aGF0CgoqIGV2ZXJ5IG9ic2VydmF0aW9uIHNob3VsZCBjb3JyZXNwb25kIHRvIGEgc2luZ2xlIHJvdzsKKiBldmVyeSB2YXJpYWJsZSBzaG91bGQgY29ycmVzcG9uZCB0byBhIHNpbmdsZSBjb2x1bW4uCgpUaWR5IGRhdGEgaXMgY29tcHV0YXRpb25hbGx5IGNvbnZlbmllbnQsIGFuZCBtYW55IG9mIHRoZSB0b29scyB3ZSB3aWxsCnVzZSBhcmUgZGVzaWduZWQgYXJvdW5kIHRpZHkgZGF0YSBmcmFtZXMuCgpBIGxhcmdlIHJhbmdlIG9mIHRoZXNlIHRvb2xzIGNhbiBiZSBhY2Nlc3NlZCBieSBsb2FkaW5nIHRoZQpgdGlkeXZlcnNlYCBwYWNrYWdlOgoKYGBge3IsIGV2YWwgPSBGQUxTRX0KbGlicmFyeSh0aWR5dmVyc2UpCmBgYAoKQnV0IGZvciBub3cgSSB3aWxsIGxvYWQgdGhlIG5lZWRlZCBwYWNrYWdlcyBpbmRpdmlkdWFsbHkuCgo8ZGl2IGNsYXNzID0gImFsZXJ0Ij4KVGhlIHRlcm0gX3RpZHlfIGlzIGEgbGl0dGxlIHVuZm9ydHVuYXRlLgoKKiBEYXRhIHRoYXQgaXMgbm90IF90aWR5XyBpc24ndCBuZWNlc3NhcmlseSBfYmFkXy4KKiBGb3IgaHVtYW4gcmVhZGluZywgYW5kICBmb3Igc29tZSBjb21wdXRhdGlvbnMsIGRhdGEgaW4gYSB3aWRlciBmb3JtYXQgY2FuCiAgYmUgYmV0dGVyLgoqIEZvciBvdGhlciBjb21wdXRhdGlvbnMgZGF0YSBpbiBhIGxvbmdlciwgb3IgbmFycm93ZXIsIGZvcm1hdCBjYW4gYmUgYmV0dGVyLgo8L2Rpdj4KCgojIyBUaWJibGVzCgpNYW55IHRvb2xzIGluIHRoZSBgdGlkeXZlcnNlYCBwcm9kdWNlIHNsaWdodGx5IGVuaGFuY2VkIGRhdGEgZnJhbWVzCmNhbGxlZCBfdGliYmxlc186CgpgYGB7cn0KbGlicmFyeSh0aWJibGUpCmZhaXRoZnVsX3RibCA8LSBhc190aWJibGUoZmFpdGhmdWwpCmNsYXNzKGZhaXRoZnVsX3RibCkKYGBgClRpYmJsZXMgcHJpbnQgZGlmZmVyZW50bHkgZnJvbSBzdGFuZGFyZCBkYXRhIGZyYW1lczoKCmBgYHtyLCBoaWdobGlnaHQub3V0cHV0PWMoMSwgMywgMTQpfQpmYWl0aGZ1bF90YmwKYGBgCgpGb3IgdGhlIG1vc3QgcGFydCBkYXRhIGZyYW1lcyBhbmQgdGliYmxlcyBjYW4gYmUgdXNlZCBpbnRlcmNoYW5nZWFibHkuCgoKIyMgVGlkeWluZyBEYXRhCgpNYW55IGRhdGEgc2V0cyBhcmUgaW4gdGlkeSBmb3JtIGFscmVhZHkuCgpJZiB0aGV5IGFyZSBub3QsIHRoZXkgY2FuIGJlIHB1dCBpbnRvIHRpZHkgZm9ybS4KClRoZSB0b29scyBmb3IgdGhpcyBhcmUgcGFydCBvZiBfZGF0YSB0ZWNobm9sb2dpZXNfLgoKVGhlIHRhc2tzIGludm9sdmVkIGFyZSBwYXJ0IG9mIHdoYXQgaXMgc29tZXRpbWVzIGNhbGxlZCBfZGF0YQp3cmFuZ2xpbmdfLgoKCiMjIEFuIEV4YW1wbGU6IEdsb2JhbCBBdmVyYWdlIFN1cmZhY2UgVGVtcGVyYXR1cmVzCgpBbW9uZyBtYW55IGRhdGEgc2V0cyBhdmFpbGFibGUgYXQKPGh0dHBzOi8vZGF0YS5naXNzLm5hc2EuZ292L2dpc3RlbXAvPiBpcyBkYXRhIG9uIG1vbnRobHkgZ2xvYmFsCmF2ZXJhZ2Ugc3VyZmFjZSB0ZW1wZXJhdHVyZXMgb3ZlciBhIG51bWJlciBvZiB5ZWFycy4KCjwhLS0gcGF5d2FsbGVkIG5vdywgc28gdXNlIHd3dy9hcmNoaXZlLm9yZyAtLT4KClRoZXNlIGRhdGEgZnJvbSAyMDE3IHdlcmUgdXNlZCBmb3IgdGhlIHdpZGVseSBjaXRlZApbQmxvb21iZXJnIGhvdHRlc3QgeWVhciB2aXN1YWxpemF0aW9uXShodHRwczovL3dlYi5hcmNoaXZlLm9yZy93ZWIvMjAxOTAyMDIxOTQ0MzIvaHR0cHM6Ly93d3cuYmxvb21iZXJnLmNvbS9ncmFwaGljcy9ob3R0ZXN0LXllYXItb24tcmVjb3JkLykuCgpUaGUgY3VycmVudCBkYXRhIGFyZSBhdmFpbGFibGUgaW4gYSBmb3JtYXR0ZWQgdGV4dCBmaWxlIGF0Cgo8aHR0cHM6Ly9kYXRhLmdpc3MubmFzYS5nb3YvZ2lzdGVtcC90YWJsZWRhdGFfdjQvR0xCLlRzK2RTU1QudHh0PgoKb3IgYXMgYQpbX0NTVl8gKGNvbW1hLXNlcGFyYXRlZCB2YWx1ZXMpXShodHRwczovL2VuLndpa2lwZWRpYS5vcmcvd2lraS9Db21tYS1zZXBhcmF0ZWRfdmFsdWVzKSBmaWxlIGF0Cgo8aHR0cHM6Ly9kYXRhLmdpc3MubmFzYS5nb3YvZ2lzdGVtcC90YWJsZWRhdGFfdjQvR0xCLlRzK2RTU1QuY3N2PgoKYGBge3IgZG93bmxvYWQtR0xCLCBlY2hvID0gRkFMU0V9CmBgYAoKVGhlIGZpcnN0IGZldyBsaW5lcyBvZiB0aGUgQ1NWIGZpbGU6CgpgYGB7cn0KI3wgZWNobzogZmFsc2UKI3wgY29tbWVudDogIiAgICIKcmVhZExpbmVzKCJHTEIuVHMrZFNTVC5jc3YiLCA2KSB8PiB3cml0ZUxpbmVzKCkKYGBgCgpUaGUgQ1NWIGZpbGUgaXMgYSBsaXR0bGUgZWFzaWVyIChmb3IgYSBjb21wdXRlciBwcm9ncmFtKSB0byByZWFkIGluLApzbyB3ZSB3aWxsIHdvcmsgd2l0aCB0aGF0LgoKVGhlIG51bWJlcnMgaW4gdGhlIENTViBmaWxlIHJlcHJlc2VudCBkZXZpYXRpb25zIGluIGRlZ3JlZXMgQ2VsY2l1cwpmcm9tIHRoZSBhdmVyYWdlIHRlbXBlcmF0dXJlIGZvciB0aGUgYmFzZSBwZXJpb2QgMTk1MS0xOTgwLgoKVGhlIGZpbGUgYXZhaWxhYmxlIG9uIEphbnVhcnkgMTYsIDIwMjUsIGlzIG5vdyBhdmFpbGFibGUKW2xvY2FsbHldKGh0dHBzOi8vc3RhdC51aW93YS5lZHUvfmx1a2UvZGF0YS9HTEIuVHMrZFNTVC5jc3YpLgoKV2UgY2FuIG1ha2Ugc3VyZSBpdCBoYXMgYmVlbiBkb3dubG9hZGVkIHRvIG91ciB3b3JraW5nIGRpcmVjdG9yeSB3aXRoCgpgYGB7ciBkb3dubG9hZC1HTEJ9CmlmICghIGZpbGUuZXhpc3RzKCJHTEIuVHMrZFNTVC5jc3YiKSkKICAgIGRvd25sb2FkLmZpbGUoImh0dHBzOi8vc3RhdC51aW93YS5lZHUvfmx1a2UvZGF0YS9HTEIuVHMrZFNTVC5jc3YiLAogICAgICAgICAgICAgICAgICAiR0xCLlRzK2RTU1QuY3N2IikKYGBgCgpBc3N1bWluZyB0aGlzIGxvY2FsbHkgYXZhaWxhYmxlIGZpbGUgaGFzIGJlZW4gZG93bmxvYWRlZCwgd2UgY2FuIHJlYWQKaW4gdGhlIGRhdGEgYW5kIGRyb3Agc29tZSBjb2x1bW5zIHdlIGRvbid0IG5lZWQgd2l0aAoKYGBge3J9CmxpYnJhcnkocmVhZHIpCmdhc3QgPC0gcmVhZF9jc3YoIkdMQi5UcytkU1NULmNzdiIsIHNraXAgPSAxKVsxIDogMTNdCmBgYAoKPGRpdiBjbGFzcyA9ICJhbGVydCI+CiogVGhlIGZ1bmN0aW9uIGByZWFkX2NzdmAgaXMgZnJvbSB0aGUgYHJlYWRyYCBwYWNrYWdlLCB3aGljaCBpcyBwYXJ0IG9mIHRoZQogIGB0aWR5dmVyc2VgLgoqIEFuIGFsdGVybmF0aXZlIGlzIHRoZSBiYXNlIFIgZnVuY3Rpb24gYHJlYWQuY3N2YC4KPC9kaXY+CgpBIGxvb2sgYXQgdGhlIGZpcnN0IGZldyBsaW5lczoKCmBgYHtyfQpoZWFkKGdhc3QsIDUpCmBgYAoKQW5kIHRoZSBsYXN0IGZldyBsaW5lczoKCmBgYHtyfQp0YWlsKGdhc3QsIDUpCmBgYAoKVGhlIGBwcmludCgpYCBtZXRob2QgZm9yIHRpYmJsZXMgYWJicmV2aWF0ZXMgdGhlIG91dHB1dC4KCkl0IGlzIG5lYXRlciBhbmQgcHJvdmlkZXMgc29tZSB1c2VmdWwgYWRkaXRpb25hbCBpbmZvcm1hdGlvbiBvbgp2YXJpYWJsZSBkYXRhIHR5cGVzLgoKQnV0IGl0IHNob3dzIG9ubHkgdGhlIGZpcnN0IGZldyByb3dzIGFuZCBtYXkgbm90IGV4cGxpY2l0bHkgc2hvdyBzb21lCmNvbHVtbnMuCgpJZiBzb21lIGNvbHVtbnMgYXJlIHNraXBwZWQsIHlvdSBjYW4gYXNrIHRvIHNlZSBtb3JlIGJ5IGNhbGxpbmcKYHByaW50KClgIGV4cGxpY2l0bHk6CgpgYGB7cn0KcHJpbnQodGFpbChnYXN0KSwgd2lkdGggPSAxMDApCmBgYAoKPCEtLQoqIFRoZSBsYXN0IHZhbHVlcyBpbiB0aGUgYEF1Z2AgLSBgRGVjYCBjb2x1bW5zIGFyZSBtaXNzaW5nIGFuZCBjb2RlZCBhcyBgKioqYC4KKiBUaGlzIGNhdXNlcyB0aGVzZSBjb2x1bW5zIHRvIGJlIHJlYWQgYXMgY2hhcmFjdGVyIHZlY3RvcnMgaW5kaWNhdGVkCiAgYnkgYDxjaHI+YC4KKiBUaGUgb3RoZXJzIGhhdmUgYmVlbiByZWFkIGFzIG51bWVyaWMsIGNvZGVkIGA8ZGJsPmAgKGZvciAKICBfZG91YmxlIHByZWNpc2lvbl8pLgoqIFdlIGNhbiBmaXggdGhlIHRoZXNlIGNvbHVtbnMgbm93IG9yIGRlYWwgd2l0aCB0aGVtIGxhdGVyLgoKT25lIHdheSB0byBmaXggdGhlbSBub3cgaXMgdG8gdXNlIGBtdXRhdGVfaWZgOgoKYGBge3IsIHdhcm5pbmcgPSBGQUxTRX0KZ2FzdCA8LSBtdXRhdGUoZ2FzdCwgYWNyb3NzKHdoZXJlKGlzLmNoYXJhY3RlciksIGFzLm51bWVyaWMpKQpoZWFkKGdhc3QpCmBgYAoKRWFjaCBvYnNlcnZhdGlvbiBjb25zaXN0cyBvZiBhIHllYXIsIGEgbW9udGgsIGFuZCBhIHRlbXBlcmF0dXJlLgotLT4KClRoZSBmb3JtYXQgd2l0aCBvbmUgY29sdW1uIHBlciBtb250aCBpcyBjb21wYWN0IGFuZCB1c2VmdWwgZm9yIHZpZXdpbmcKYW5kIGRhdGEgZW50cnkuCgpCdXQgaXQgaXMgbm90IGluIF90aWR5IGZvcm1hdF8gc2luY2UKCiogdGhlIG1vbnRobHkgdGVtcGVyYXR1cmUgIHZhcmlhYmxlIGlzIHNwcmVhZCBvdmVyIDEyIGNvbHVtbnM7CiogdGhlIG1vbnRoIHZhcmlhYmxlIGlzIGVuY29kZWQgaW4gdGhlIGNvbHVtbiBoZWFkaW5ncy4KCkZvciBvYnZpb3VzIHJlYXNvbnMgdGhpcyBkYXRhIGZvcm1hdCBpcyBvZnRlbiByZWZlcnJlZCB0byBhcyBfd2lkZQpmb3JtYXRfLgoKVGhlIHRpZHksIG9yIF9sb25nXywgZm9ybWF0IHdvdWxkIGhhdmUgdGhyZWUgdmFyaWFibGVzOiBgWWVhcmAsCmBNb250aGAsIGFuZCBgVGVtcGAuCgpPbmUgd2F5IHRvIHB1dCB0aGlzIGRhdGEgZnJhbWUgaW4gdGlkeSwgb3IgbG9uZywgZm9ybWF0IHVzZXMKYHBpdm90X2xvbmdlcigpYCBmcm9tIHRoZSBgdGlkeXJgIHBhY2thZ2U6CgpgYGB7cn0KbGlicmFyeSh0aWR5cikKbGdhc3QgPC0gcGl2b3RfbG9uZ2VyKGdhc3QsCiAgICAgICAgICAgICAgICAgICAgICAtWWVhciwgICMjIHNwZWNpZmllcyB0aGUgY29sdW1ucyB0byB1c2UgLS0gYWxsIGJ1dCBZZWFyCiAgICAgICAgICAgICAgICAgICAgICBuYW1lc190byA9ICJNb250aCIsCiAgICAgICAgICAgICAgICAgICAgICB2YWx1ZXNfdG8gPSAiVGVtcCIpCmBgYAoKVGhlIGZpcnN0IGZldyByb3dzIG9mIHRoZSByZXN1bHQ6CgpgYGB7cn0KaGVhZChsZ2FzdCkKYGBgCgpEdXJpbmcgcGxvdHRpbmcgaXQgaXMgbGlrZWx5IHRoYXQgdGhlIGBNb250aGAgdmFyaWFibGUgd2lsbCBiZQpjb252ZXJ0ZWQgdG8gYSBfZmFjdG9yXy4KCkJ5IGRlZmF1bHQsIHRoaXMgd2lsbCBvcmRlciBsZXZlbHMgYWxwaGFiZXRpY2FsbHksIHdoaWNoIGlzIG5vdCB3aGF0IHdlIHdhbnQ6CgpgYGB7cn0KbGV2ZWxzKGFzLmZhY3RvcihsZ2FzdCRNb250aCkpCmBgYAoKV2UgY2FuIGd1YXJkIGFnYWluc3QgdGhpcyBieSBjb252ZXJ0aW5nIGBNb250aGAgdG8gYSBmYWN0b3Igd2l0aCB0aGUKcmlnaHQgbGV2ZWxzIG5vdzoKCmBgYHtyfQpsZ2FzdCA8LSBtdXRhdGUobGdhc3QsIE1vbnRoID0gZmFjdG9yKE1vbnRoLCBsZXZlbHMgPSBtb250aC5hYmIpKQpsZXZlbHMobGdhc3QkTW9udGgpCmBgYAoKPCEtLSBwYXl3YWxsZWQgbm93LCBzbyB1c2Ugd3d3L2FyY2hpdmUub3JnIC0tPgoKV2UgY2FuIG5vdyB1c2UgdGhpcyB0aWR5IHZlcnNpb24gb2YgdGhlIGRhdGEgdG8gY3JlYXRlIGEgc3RhdGljCnZlcnNpb24gb2YgdGhlIFtCbG9vbWJlcmcgaG90dGVzdCB5ZWFyCnZpc3VhbGl6YXRpb25dKGh0dHBzOi8vd2ViLmFyY2hpdmUub3JnL3dlYi8yMDE5MDIwMjE5NDQzMi9odHRwczovL3d3dy5ibG9vbWJlcmcuY29tL2dyYXBoaWNzL2hvdHRlc3QteWVhci1vbi1yZWNvcmQvKS4KClRoZSBiYXNpYyBmcmFtZXdvcmsgaXMgc2V0IHVwIHdpdGgKCmBgYHtyfQpsaWJyYXJ5KGdncGxvdDIpCnAgPC0gZ2dwbG90KGxnYXN0KSArCiAgICBnZ3RpdGxlKCJHbG9iYWwgQXZlcmFnZSBTdXJmYWNlIFRlbXBlcmF0dXJlcyIpICsKICAgIHRoZW1lKHBsb3QudGl0bGUgPSBlbGVtZW50X3RleHQoaGp1c3QgPSAwLjUpKQpgYGAKCjxkaXYgY2xhc3MgPSAiYWxlcnQiPgpgZ2dwbG90YCBvYmplY3RzIG9ubHkgcHJvZHVjZSBvdXRwdXQgd2hlbiB0aGV5IGFyZSBwcmludGVkLgoKVG8gc2VlIHRoZSBwbG90IGluIGBwYCB3ZSBuZWVkIHRvIHByaW50IGl0LCBmb3IgZXhhbXBsZSBieSB1c2luZyBhCmxpbmUgd2l0aCBvbmx5IGBwYC4KPC9kaXY+CgpgYGB7ciBnYXN0LWJhc2UsIGV2YWwgPSBGQUxTRX0KIHAKYGBgCmBgYHtyIGdhc3QtYmFzZSwgZXZhbCA9IFRSVUUsIGVjaG8gPSBGQUxTRX0KYGBgCgpUaGVuIGFkZCBhIGxheWVyIHdpdGggbGluZXMgZm9yIGVhY2ggeWVhciAoc3BlY2lmaWVkIGJ5IHRoZSBgZ3JvdXBgCmFyZ3VtZW50IHRvIGBnZW9tX2xpbmVgKS4KCmBgYHtyIGdhc3QtbGluZXMsIGV2YWwgPSBGQUxTRX0KcCArIGdlb21fbGluZShhZXMoeCA9IE1vbnRoLAogICAgICAgICAgICAgICAgICB5ID0gVGVtcCwKICAgICAgICAgICAgICAgICAgZ3JvdXAgPSBZZWFyKSkKYGBgCmBgYHtyIGdhc3QtbGluZXMsIGV2YWwgPSBUUlVFLCBlY2hvID0gRkFMU0UsIHdhcm5pbmcgPSBGQUxTRX0KYGBgCgpXZSBjYW4gdXNlIGNvbG9yIHRvIGRpc3Rpbmdpc2ggdGhlIHllYXJzLgoKYGBge3IgZ2FzdC1jb2xvcjEsIGV2YWwgPSBGQUxTRX0KcDEgPC0gcCArCiAgICBnZW9tX2xpbmUoYWVzKHggPSBNb250aCwKICAgICAgICAgICAgICAgICAgeSA9IFRlbXAsCiAgICAgICAgICAgICAgICAgIGNvbG9yID0gWWVhciwKICAgICAgICAgICAgICAgICAgZ3JvdXAgPSBZZWFyKSwKICAgICAgICAgICAgICBuYS5ybSA9IFRSVUUpCnAxCmBgYApTYXZpbmcgdGhlIHBsb3Qgc3BlY2lmaWNhdGlvbiBpbiB0aGUgdmFyaWFibGUgYHAxYCB3aWxsIG1ha2UgaXQgZWFzaWVyCnRvIGV4cGVyaW1lbnQgd2l0aCBjb2xvciB2YXJpYXRpb25zOgoKYGBge3IgZ2FzdC1jb2xvcjEsIGV2YWwgPSBUUlVFLCBlY2hvID0gRkFMU0V9CmBgYAoKYGBge3IsIGluY2x1ZGUgPSBGQUxTRX0KIyMgQ29tcHV0ZSB0aGUgcGFzdCB5ZWFyIGFuZCBtYWtlIHN1cmUgaXQgaXMgaW4gdGhlIGZpbGUKbGlicmFyeShsdWJyaWRhdGUpCnBhc3RfeWVhciA8LSB5ZWFyKHRvZGF5KCkpIC0gMQpwYXN0X3llYXIKc3RvcGlmbm90KHBhc3RfeWVhciAlaW4lIGxnYXN0JFllYXIpCmBgYAoKT25lIHdheSB0byBoaWdobGlnaHQgdGhlIGBwYXN0X3llYXJgIGByIHBhc3RfeWVhcmA6CgpgYGB7ciBnYXN0LWNvbG9yMiwgZXZhbCA9IEZBTFNFfQpsZ2FzdF9sYXN0IDwtIGZpbHRlcihsZ2FzdCwgWWVhciA9PSBwYXN0X3llYXIpCnAxICsgZ2VvbV9saW5lKGFlcyh4ID0gTW9udGgsCiAgICAgICAgICAgICAgICAgICB5ID0gVGVtcCwKICAgICAgICAgICAgICAgICAgIGdyb3VwID0gWWVhciksCiAgICAgICAgICAgICAgIGxpbmV3aWR0aCA9IDEsCiAgICAgICAgICAgICAgIGNvbG9yID0gInJlZCIsCiAgICAgICAgICAgICAgIGRhdGEgPSBsZ2FzdF9sYXN0LAogICAgICAgICAgICAgICBuYS5ybSA9IFRSVUUpCmBgYApgYGB7ciBnYXN0LWNvbG9yMiwgZWNobyA9IEZBTFNFfQpgYGAKCkEgdXNlZnVsIHdheSB0byBzaG93IG1vcmUgcmVjZW50IGRhdGEgaW4gdGhlIGNvbnRleHQgb2YgdGhlIGZ1bGwgZGF0YQppcyB0byBzaG93IHRoZSBmdWxsIGRhdGEgaW4gZ3JleSBhbmQgdGhlIG1vcmUgcmVjZW50IHllYXJzIGluIGJsYWNrOgoKYGBge3IgZ2FzdC1mdWxsLWdyZXksIGV2YWwgPSBGQUxTRX0KbGdhc3QyayA8LSBmaWx0ZXIobGdhc3QsIFllYXIgPj0gMjAwMCkKZ2dwbG90KGxnYXN0LCBhZXMoeCA9IE1vbnRoLAogICAgICAgICAgICAgICAgICB5ID0gVGVtcCwKICAgICAgICAgICAgICAgICAgZ3JvdXAgPSBZZWFyKSkgKwogICAgZ2VvbV9saW5lKGNvbG9yID0gImdyZXk4MCIpICsKICAgIHRoZW1lX21pbmltYWwoKSArCiAgICBnZW9tX2xpbmUoZGF0YSA9IGxnYXN0MmspCmBgYApgYGB7ciBnYXN0LWZ1bGwtZ3JleSwgZWNobyA9IEZBTFNFLCB3YXJuaW5nID0gRkFMU0V9CmBgYAoKSWYgeW91IHdhbnQgdG8gdXBkYXRlIHlvdXIgcGxvdCBsYXRlciBpbiB0aGUgeWVhciB0aGVuIHRoZSBjdXJyZW50CnllYXIncyBlbnRyeSBtYXkgY29udGFpbiBtaXNzaW5nIHZhbHVlIGluZGljYXRvcnMgdGhhdCB5b3Ugd2lsbCBoYXZlCnRvIGRlYWwgd2l0aC4KPCEtLQpUaGUgdmVyc2lvbiBvZiB0aGUgZGF0YSBbb24gdGhlCndlYl0oaHR0cHM6Ly9kYXRhLmdpc3MubmFzYS5nb3YvZ2lzdGVtcC90YWJsZWRhdGFfdjMvR0xCLlRzK2RTU1QudHh0KQptYXkgaGF2ZSBiZWVuIHVwZGF0ZWQgdG8gaW5jbHVkZSB0aGUgbWlzc2luZyB2YWx1ZXMgZm9yIGByIHBhc3RfeWVhcmAuCklmIHlvdSB3YW50IHRvIHVwZGF0ZSB5b3VyIHBsb3QgbGF0ZXIgaW4gdGhlIHllYXIgeW91IHdpbGwgc2VlCnNpbWlsYXIgbWlzc2luZyB2YWx1ZSBtYXJrZXJzIGZvciB0aGUgcmVtYWluaW5nIG1vbnRocyBvZgpgciAocGFzdF95ZWFyICsgMSlgLgotLT4KClRoZSBOZXcgWW9yayBUaW1lcyBvbiBKYW51YXJ5IDE4LCAyMDE4LCBwdWJsaXNoZWQKW2Fub3RoZXIgdmlzdWFsaXphdGlvbl0oaHR0cHM6Ly93d3cubnl0aW1lcy5jb20vaW50ZXJhY3RpdmUvMjAxOC8wMS8xOC9jbGltYXRlL2hvdHRlc3QteWVhci0yMDE3Lmh0bWwpCm9mIHRoZXNlIGRhdGEgc2hvd2luZyBhdmVyYWdlIHllYXJseSB0ZW1wZXJhdHVyZXMgKFt2aWEgR29vZ2xlIG1heSB3b3JrIGJldHRlcl0oaHR0cHM6Ly93d3cuZ29vZ2xlLmNvbS91cmw/c2E9dCZyY3Q9aiZxPSZlc3JjPXMmc291cmNlPXdlYiZjZD0mY2FkPXJqYSZ1YWN0PTgmdmVkPTJhaFVLRXdqXzRQbWhuTTMxQWhYRmpZa0VIZlF4QVRFUUZub0VDQVlRQVEmdXJsPWh0dHBzJTNBJTJGJTJGd3d3Lm55dGltZXMuY29tJTJGaW50ZXJhY3RpdmUlMkYyMDE4JTJGMDElMkYxOCUyRmNsaW1hdGUlMkZob3R0ZXN0LXllYXItMjAxNy5odG1sJnVzZz1BT3ZWYXczcW91eEhRQUpDcXJ0X0YzZmJMRUdFKSkuCgpUbyByZWNyZWF0ZSB0aGlzIHBsb3Qgd2UgZmlyc3QgbmVlZCB0byBjb21wdXRlIHllYXJseSBhdmVyYWdlIHRlbXBlcmF0dXJlcy4KClRoaXMgaXMgZWFzeSB0byBkbyB3aXRoIHRoZSBgc3VtbWFyaXplKClgIGFuZCBgZ3JvdXBfYnkoKWAgZnVuY3Rpb25zCmZyb20gYGRweXJgOgoKYGBge3J9CmxpYnJhcnkoZHBseXIpCmF0ZW1wIDwtIGxnYXN0IHw+CiAgICBncm91cF9ieShZZWFyKSB8PgogICAgc3VtbWFyaXplKEF2ZVRlbXAgPSBtZWFuKFRlbXAsIG5hLnJtID0gVFJVRSkpCmhlYWQoYXRlbXApCmBgYAoKVXNpbmcgYG5hLnJtID0gVFJVRWAgZW5zdXJlcyB0aGF0IHRoZSBtZWFuIGlzIGJhc2VkIG9uIHRoZSBhdmFpbGFibGUKbW9udGhzIGlmIGRhdGEgZm9yIHNvbWUgbW9udGhzIGlzIG1pc3NpbmcuCgpBIHNpbXBsZSB2ZXJzaW9uIG9mIHRoZSBwbG90IGlzIHRoZW4gcHJvZHVjZWQgYnkKCmBgYHtyIGdhc3Qtbnl0LCBldmFsID0gRkFMU0V9CmdncGxvdChhdGVtcCkgKwogICAgZ2VvbV9wb2ludChhZXMoeCA9IFllYXIsIHkgPSBBdmVUZW1wKSkKYGBgCmBgYHtyIGdhc3Qtbnl0LCBlY2hvID0gRkFMU0V9CmBgYAoKQSB2YXJpYXRpb24gc2hvd2luZyByZWNvcmQgeWVhcnM6CgpgYGB7ciAgZ2FzdC1ueXQtcmVjLCBldmFsID0gRkFMU0V9CmxpYnJhcnkoZ2dyZXBlbCkKYXRlbXBfcmVjIDwtIGZpbHRlcihhdGVtcCwgY3VtbWF4KEF2ZVRlbXApID09IEF2ZVRlbXApCmdncGxvdChhdGVtcCwgYWVzKHggPSBZZWFyLCB5ID0gQXZlVGVtcCkpICsKICAgIGdlb21fcG9pbnQoKSArCiAgICBnZW9tX3BvaW50KGRhdGEgPSBhdGVtcF9yZWMsIGNvbG9yID0gInJlZCIpICsKICAgIGdlb21fdGV4dF9yZXBlbChhZXMobGFiZWwgPSBZZWFyKSwKICAgICAgICAgICAgICAgICAgICBkYXRhID0gYXRlbXBfcmVjLAogICAgICAgICAgICAgICAgICAgIGNvbG9yID0gImJsdWUiKQpgYGAKYGBge3IgZ2FzdC1ueXQtcmVjLCBlY2hvID0gRkFMU0V9CmBgYAoKQW5vdGhlciB2YXJpYXRpb24gb24gdGhlIEJsb29tYmVyZyBwbG90IHNob3dpbmcganVzdCBhIGZldyB5ZWFycyAyMAp5ZWFycyBhcGFydDoKCmBgYHtyIGdhc3Qtc2tpcCwgZXZhbCA9IEZBTFNFfQpsZ19ieV8yMCA8LQogICAgZmlsdGVyKGxnYXN0LAogICAgICAgICAgIFllYXIgJWluJSBzZXEoMjAyMCwgYnkgPSAtMjAsIGxlbiA9IDUpKSB8PgogICAgbXV0YXRlKFllYXIgPSBmYWN0b3IoWWVhcikpCmdncGxvdChsZ19ieV8yMCwgYWVzKHggPSBNb250aCwKICAgICAgICAgICAgICAgICAgICAgeSA9IFRlbXAsCiAgICAgICAgICAgICAgICAgICAgIGdyb3VwID0gWWVhciwKICAgICAgICAgICAgICAgICAgICAgY29sb3IgPSBZZWFyKSkgKwogICAgZ2VvbV9saW5lKCkKYGBgCkNvbnZlcnRpbmcgYFllYXJgIHRvIGEgZmFjdG9yIHJlc3VsdHMgaW4gYSBkaXNjcmV0ZSBjb2xvciBzY2FsZSBhbmQgbGVnZW5kLgpgYGB7ciBnYXN0LXNraXAsIGVjaG8gPSBGQUxTRX0KYGBgCgoKIyMgSGFuZGxpbmcgTWlzc2luZyBWYWx1ZXMKClRoZSBkYXRhIGZvciAyMDE5IGF2YWlsYWJsZSBpbiBlYXJseSAyMDIwIGlzIGFsc28gYXZhaWxhYmxlCltsb2NhbGx5XShodHRwczovL3N0YXQudWlvd2EuZWR1L35sdWtlL2RhdGEvR0xCLlRzK2RTU1QtMjAxOS5jc3YpLgoKYGBge3IsIGVjaG8gPSBGQUxTRX0KaWYgKCEgZmlsZS5leGlzdHMoIkdMQi5UcytkU1NULTIwMTkuY3N2IikpCiAgICBkb3dubG9hZC5maWxlKCJodHRwczovL3N0YXQudWlvd2EuZWR1L35sdWtlL2RhdGEvR0xCLlRzK2RTU1QtMjAxOS5jc3YiLAogICAgICAgICAgICAgICAgICAiR0xCLlRzK2RTU1QtMjAxOS5jc3YiKQpgYGAKCkFzc3VtaW5nIHRoaXMgbG9jYWxseSBhdmFpbGFibGUgZmlsZSBoYXMgYmVlbiBkb3dubG9hZGVkLCB3ZSBjYW4gcmVhZAppbiB0aGUgZGF0YSBhbmQgZHJvcCBzb21lIGNvbHVtbnMgd2UgZG9uJ3QgbmVlZCB3aXRoCgpgYGB7cn0KZ2FzdDIwMTkgPC0gcmVhZF9jc3YoIkdMQi5UcytkU1NULTIwMTkuY3N2Iiwgc2tpcCA9IDEpWzEgOiAxM10KYGBgCgpUaGUgbGFzdCB0aHJlZSBjb2x1bW5zIGFyZSByZWFkIGFzIGNoYXJhY3RlciB2YXJpYWJsZXM6CgpgYGB7cn0KaGVhZChnYXN0MjAxOSwgNSkKYGBgCgpUaGUgcmVhc29uIGlzIHRoYXQgZGF0YSBmb3IgT2N0b2JlciB0aHJvdWdoIERlY2VtYmVyIHdlcmUgbm90CmF2YWlsYWJsZToKCmBgYHtyfQp0YWlsKGdhc3QyMDE5LCAyKQpgYGAKCldlIHdhbnQgdG8gY29udmVydCB0aGVzZSBjb2x1bW5zIHRvIG51bWVyaWMsIHdpdGggbWlzc2luZyB2YWx1ZXMKcmVwcmVzZW50ZWQgYXMgYE5BYC4KCk9uZSBvcHRpb24gaXMgdG8gaGFuZGxlIHRoZW0gaW5kaXZpZHVhbGx5OgoKYGBge3J9Cmdhc3QyMDE5JE9jdCA8LSBhcy5udW1lcmljKGdhc3QyMDE5JE9jdCkKdGFpbChnYXN0MjAxOSwgMikKYGBgCgpBbm90aGVyIG9wdGlvbiBpcyB0byBjb252ZXJ0IGFsbCBjaGFyYWN0ZXIgY29sdW1ucyB0byBudW1lcmljIHdpdGgKCmBgYHtyfQpnYXN0MjAxOSA8LSBtdXRhdGUoZ2FzdDIwMTksIGFjcm9zcyh3aGVyZShpcy5jaGFyYWN0ZXIpLCBhcy5udW1lcmljKSkKdGFpbChnYXN0MjAxOSwgMikKYGBgCgpUaGUgd2FybmluZ3MgYXJlIGJlbmlnbiBhbmQgY2FuIGJlIHN1cHByZXNzZWQgd2l0aCB0aGUgYHdhcm5pbmcgPQpGQUxTRWAgY2h1bmsgb3B0aW9uLgoKU2luY2Ugd2Uga25vdyB0aGUgbWlzc2luZyB2YWx1ZSBwYXR0ZXJuIGAqKipgIHdlIGNhbiBhbHNvIGF2b2lkIHRoZQpuZWVkIHRvIGZpeCB0aGUgZGF0YSBhZnRlciB0aGUgZmFjdCBieSBzcGVjaWZ5aW5nIHRoaXMgYXQgcmVhZCB0aW1lOgoKYGBge3J9Cmdhc3QyMDE5IDwtIHJlYWRfY3N2KCJHTEIuVHMrZFNTVC0yMDE5LmNzdiIsIG5hID0gIioqKiIsIHNraXAgPSAxKVsxIDogMTNdCnRhaWwoZ2FzdDIwMTksIDIpCmBgYAoKQSBwbG90IGhpZ2hsaWdodGluZyB0aGUgeWVhciAyMDE5IHNob3dzIG9ubHkgdGhlIG1vbnRocyB3aXRoIGF2YWlsYWJsZQpkYXRhOgoKYGBge3IgZ2FzdC0yMDE5LCBldmFsID0gRkFMU0V9CmxnYXN0MjAxOSA8LSBnYXN0MjAxOSB8PgogICAgcGl2b3RfbG9uZ2VyKC1ZZWFyLAogICAgICAgICAgICAgICAgIG5hbWVzX3RvID0gIk1vbnRoIiwKICAgICAgICAgICAgICAgICB2YWx1ZXNfdG8gPSAiVGVtcCIpIHw+CiAgICBtdXRhdGUoTW9udGggPSBmYWN0b3IoTW9udGgsIGxldmVscyA9IG1vbnRoLmFiYikpCmdncGxvdChsZ2FzdDIwMTksIGFlcyh4ID0gTW9udGgsCiAgICAgICAgICAgICAgICAgICAgICB5ID0gVGVtcCwKICAgICAgICAgICAgICAgICAgICAgIGdyb3VwID0gWWVhcikpICsKICAgIGdlb21fbGluZShjb2xvciA9ICJncmV5ODAiLAogICAgICAgICAgICAgIG5hLnJtID0gVFJVRSkgKwogICAgZ2VvbV9saW5lKGRhdGEgPSBmaWx0ZXIobGdhc3QyMDE5LCBZZWFyID09IDIwMTkpLAogICAgICAgICAgICAgIG5hLnJtID0gVFJVRSkKYGBgCgpBZGRpbmcgYG5hLnJtID0gVFJVRWAgaW4gdGhlIGBnZW9tX2xpbmVgIGNhbGxzIHN1cHByZXNzZXMgd2FybmluZ3M7CnRoZSBwbG90IHdvdWxkIGJlIHRoZSBzYW1lIHdpdGhvdXQgdGhlc2UuCgpgYGB7ciBnYXN0LTIwMTksIGVjaG8gPSBGQUxTRX0KYGBgCgo8ZGl2IGNsYXNzID0gImFsZXJ0Ij4KT3V0bGluZSBvZiB0aGUgdG9vbHMgdXNlZDoKCiogRGF0YSBwcm9jZXNzaW5nOgogICAgKiBSZWFkaW5nOiBgcmVhZF9jc3ZgLCBgcmVhZC5jc3ZgOwogICAgKiBSZXNoYXBpbmc6IGBwaXZvdF9sb25nZXJgOwogICAgKiBDbGVhbmluZzogYGlzLmNoYXJhY3RlcmAsIGBhcy5udW1lcmljYCwgYG11dGF0ZWAsIGBhY3Jvc3NgLCBgZmFjdG9yYDsKICAgICogU3VtbWFyaXppbmc6IGBncm91cF9ieWAsIGBzdW1tYXJpemVgLgoqIFZpc3VhbGl6YXRpb24gZ2VvbWV0cmllczoKICAgICogYGdlb21fbGluZWAgZm9yIGEgbGluZSBwbG90OwogICAgKiBgZ2VvbV9wb2ludGAgZm9yIGEgc2NhdHRlciBwbG90Lgo8L2Rpdj4KCgojIyBSZWFkaW5nCgpTdGV2ZW5zJyBjbGFzc2lmaWNhdGlvbiBvZiBzY2FsZXMgb2YgbWVhc3VyZW1lbnQgaXMgZGVzY3JpYmVkIGluIGEKW1dpa2lwZWRpYQphcnRpY2xlXShodHRwczovL2VuLndpa2lwZWRpYS5vcmcvd2lraS9MZXZlbF9vZl9tZWFzdXJlbWVudCkuCgpBIGdvb2QgaW50cm9kdWN0aW9uIHRvIHRoZSBjb25jZXB0IG9mIF90aWR5IGRhdGFfIGlzIHByb3ZpZGVkIGluIGEKW2NoYXB0ZXIgaW4gX1IgZm9yIERhdGEKU2NpZW5jZV9dKGh0dHBzOi8vcjRkcy5oYWRsZXkubnovZGF0YS10aWR5Lmh0bWwpLgoKCiMjIEludGVyYWN0aXZlIFR1dG9yaWFsCgpBbiBpbnRlcmFjdGl2ZSBbYGxlYXJucmBdKGh0dHBzOi8vcnN0dWRpby5naXRodWIuaW8vbGVhcm5yLykgdHV0b3JpYWwKZm9yIHRoZXNlIG5vdGVzIGlzIFthdmFpbGFibGVdKGByIFdMTksoInR1dG9yaWFscy9kYXRhZnJtLlJtZCIpYCkuCgpZb3UgY2FuIHJ1biB0aGUgdHV0b3JpYWwgd2l0aAoKYGBge3IsIGV2YWwgPSBGQUxTRX0KU1RBVDQ1ODA6OnJ1blR1dG9yaWFsKCJkYXRhZnJtIikKYGBgCgoKIyMgRXhlcmNpc2VzCgoxLiBXaGljaCBvZiB0aGUgU3RldmVucyBjbGFzc2lmaWNhdGlvbnMgKG5vbWluYWwsIG9yZGluYWwsIGludGVydmFsLCByYXRpbykKICAgYmVzdCBjaGFyYWN0ZXJpemVzIHRoZXNlIHZhcmlhYmxlczoKCiAgICBhLiBEYWlseSBtYXhpbWFsIHRlbXBlcmF0dXJlcyBpbiBJb3dhIENpdHkuCiAgICBiLiBQb3B1bGF0aW9uIGNvdW50cyBmb3IgSW93YSBjb3VudGllcy4KICAgIGMuIEVkdWNhdGlvbiBsZXZlbCBvZiBqb2IgYXBwbGljYW50cyB1c2luZyB0aGUgW0J1cmVhdSBvZiBMYWJvcgogICAgICAgU3RhdGlzdGljcwogICAgICAgY2xhc3NpZmljYXRpb25dKGh0dHBzOi8vd3d3LmJscy5nb3YvY2FyZWVyb3V0bG9vay8yMDE0L2FydGljbGUvZWR1Y2F0aW9uLWxldmVsLWFuZC1qb2JzLmh0bSkuCiAgICBkLiBNYWpvciBvZiBVSSBzdHVkZW50cy4KCjwhLS0KV2hpY2ggYW5zd2VycyBhcmUgY29ycmVjdCBmb3IgZXhlcmNpc2UgMToKKiBhOiBpbnRlcnZhbDsgYjogcmF0aW87IGM6IG9yZGluYWw7IGQ6IG5vbWluYWwKICBhOiBub21pbmFsOyBiOiBpbnRlcnZhbDsgYzogb3JkaW5hbDsgZDogcmF0aW8KICBhOiBub21pbmFsOyBiOiBpbnRlcnZhbDsgYzogb3JkaW5hbDsgZDogcmF0aW8KICBhOiBpbnRlcnZhbDsgYjogcmF0aW87IGM6IG5vbWluYWw7IGQ6IG9yZGluYWwKZm10IDwtIGZ1bmN0aW9uKHgpCiAgICBwYXN0ZShsZXR0ZXJzW3NlcV9hbG9uZyh4KV0sIHgsIHNlcCA9ICI6ICIsIGNvbGxhcHNlID0gIjsgIikKc3RldiA8LSBjKCJub21pbmFsIiwgIm9yZGluYWwiLCAiaW50ZXJ2YWwiLCAicmF0aW8iKQpmbXQoc2FtcGxlKHN0ZXYsIDQpKQotLT4KCjIuIFdoaWNoIG9mIHRoZXNlIGRhdGEgc2V0cyBhcmUgaW4gdGlkeSBmb3JtPwoKICAgIGEuIFRoZSBidWlsdGluIGRhdGEgc2V0IGBjbzJgCiAgICBiLiBUaGUgYnVpbHRpbiBkYXRhIHNldCBgQk9EYAogICAgYy4gVGhlIGB3aG9gIGRhdGEgc2V0IGluIHBhY2thZ2UgYHRpZHlyYCAoYHRpZHlyOjp3aG9gKQogICAgZC4gVGhlIGBtcGdgIGRhdGEgc2V0IGluIHBhY2thZ2UgYGdncGxvdDJgIChgZ2dwbG90Mjo6bXBnYCkKCjwhLS0KV2hpY2ggYW5zd2VycyBhcmUgY29ycmVjdCBmb3IgZXhlcmNpc2UgMjoKKiBhOiBub3QgdGlkeTsgYjogdGlkeTsgYzogbm90IHRpZHk7IGQ6IHRpZHkKKiBhOiB0aWR5OyBiOiB0aWR5OyBjOiBub3QgdGlkeTsgZDogdGlkeQoqIGE6IG5vdCB0aWR5OyBiOiBub3QgdGlkeTsgYzogbm90IHRpZHk7IGQ6IHRpZHkKKiBhOiBub3QgdGlkeTsgYjogdGlkeTsgYzogdGlkeTsgZDogbm90IHRpZHkKLS0+CgpUaGUgbmV4dCBleGVyY2lzZXMgdXNlIHRoZSBkYXRhIGluIHRoZSB2YXJpYWJsZSBgZ2FwbWluZGVyYCBpbiB0aGUgcGFja2FnZQpgZ2FwbWluZGVyYC4gWW91IGNhbiBtYWtlIGl0IGF2YWlsYWJsZSB3aXRoCgpgYGB7ciwgZXZhbCA9IEZBTFNFfQpkYXRhKGdhcG1pbmRlciwgcGFja2FnZSA9ICJnYXBtaW5kZXIiKQpgYGAKMy4gVXNlIHRoZSBmdW5jdGlvbiBgc3RyYCB0byBleGFtaW5lIHRoZSB2YWx1ZSBvZiB0aGUgZ2FwbWluZGVyCiAgIHZhcmlhYmxlLiAgSG93IG1hbnkgY2FzZXMgYXJlIHRoZXJlIGluIHRoZSBkYXRhIHNldD8gSG93IG1hbnkgb2YKICAgdGhlIHZhcmlhYmxlcyBhcmUgZmFjdG9ycz8KCjQuIFVzZSB0aGUgZnVuY3Rpb25zIGBjbGFzc2AgYW5kIGBuYW1lc2AgdG8gZmluZCB0aGUgY2xhc3MgYW5kCiAgIHZhcmlhYmxlIG5hbWVzIGluIHRoZSBgZ2FwbWluZGVyYCBkYXRhLgoKNS4gVXNlIGBzdW1tYXJ5YCB0byBjb21wdXRlIHN1bW1hcnkgaW5mb3JtYXRpb24gZm9yIHRoZSB2YXJpYWJsZXMuCgo2LiBGaWxsIGluIHRoZSB2YWx1ZXMgZm9yIGAtLS1gIG5lZWRlZCB0byBwcm9kdWNlIHBsb3RzIG9mIGxpZmUKICAgZXhwZWN0YW5jeSBhZ2FpbnN0IHllYXIgZm9yIHRoZSBjb3VudHJpZXMgaW4gY29udGluZW50IE9jZWFuaWEuCjwhLS0gIyMgbm9saW50IHN0YXJ0IC0tPgpgYGB7ciwgZXZhbCA9IEZBTFNFfQpsaWJyYXJ5KGRwbHlyKQpsaWJyYXJ5KGdncGxvdDIpCmRhdGEoZ2FwbWluZGVyLCBwYWNrYWdlID0gImdhcG1pbmRlciIpCmdncGxvdChmaWx0ZXIoZ2FwbWluZGVyLCBjb250aW5lbnQgPT0gIk9jZWFuaWEiKSwKICAgICAgIGFlcyh4ID0gLS0tLCB5ID0gLS0tLCBjb2xvciA9IGNvdW50cnkpKSArCiAgICBnZW9tX2xpbmUoKQpgYGAKPCEtLSAjIyBub2xpbnQgZW5kIC0tPgo=