Visualizing a Categorical Variable
Categorical Data
Categorical data can be
- nominal, qualitative
- ordinal
For visualization, the main difference is that ordinal data suggests a particular display order.
Purely categorical data can come in a range of formats. The most common are
- raw data: individual observations;
- aggregated data: counts for each unique combination of levels
- cross-tabulated data
Raw Data
Raw data for a survey of individuals that records hair color, eye color, and gender of 592 individuals might look like this:
head(raw)
## Hair Eye Sex
## 1 Brown Blue Male
## 2 Brown Brown Male
## 3 Brown Hazel Male
## 4 Blond Green Female
## 5 Brown Brown Female
## 6 Brown Hazel MaleAggregated Data
One way to aggregate raw categorical data is to use count from dplyr:
library(dplyr)
agg <- count(raw, Hair, Eye, Sex)
head(agg)
## Hair Eye Sex n
## 1 Black Brown Male 32
## 2 Black Brown Female 36
## 3 Black Blue Male 11
## 4 Black Blue Female 9
## 5 Black Hazel Male 10
## 6 Black Hazel Female 5The count_ function from dplyr allows the variables to use to be read from the data:
agg <- count_(raw, names(raw))
## Warning: `count_()` was deprecated in dplyr 0.7.0.
## Please use `count()` instead.
## See vignette('programming') for more help
## This warning is displayed once every 8 hours.
## Call `lifecycle::last_lifecycle_warnings()` to see where this warning was generated.
head(agg)
## Hair Eye Sex n
## 1 Black Brown Male 32
## 2 Black Brown Female 36
## 3 Black Blue Male 11
## 4 Black Blue Female 9
## 5 Black Hazel Male 10
## 6 Black Hazel Female 5Cross-Tabulated Data
Cross-tabulated data can be produced from aggregate data using xtabs:
xtabs(n ~ Hair + Eye + Sex, data = agg)
## , , Sex = Male
##
## Eye
## Hair Brown Blue Hazel Green
## Black 32 11 10 3
## Brown 53 50 25 15
## Red 10 10 7 7
## Blond 3 30 5 8
##
## , , Sex = Female
##
## Eye
## Hair Brown Blue Hazel Green
## Black 36 9 5 2
## Brown 66 34 29 14
## Red 16 7 7 7
## Blond 4 64 5 8Cross-tabulated data can be produced from raw data using table:
xtb <- table(raw)
xtb
## , , Sex = Male
##
## Eye
## Hair Brown Blue Hazel Green
## Black 32 11 10 3
## Brown 53 50 25 15
## Red 10 10 7 7
## Blond 3 30 5 8
##
## , , Sex = Female
##
## Eye
## Hair Brown Blue Hazel Green
## Black 36 9 5 2
## Brown 66 34 29 14
## Red 16 7 7 7
## Blond 4 64 5 8Both raw and aggregate date in this example are in tidy form; the cross-tabulated date is not.
Cross-tabulated data on \(p\) variables is arranged in a \(p\)-way array.
The cross-tabulated data can be converted to the tidy aggregate form using
as.data.frame:
class(xtb)
## [1] "table"
head(as.data.frame(xtb))
## Hair Eye Sex Freq
## 1 Black Brown Male 32
## 2 Brown Brown Male 53
## 3 Red Brown Male 10
## 4 Blond Brown Male 3
## 5 Black Blue Male 11
## 6 Brown Blue Male 50The variable xtb corresponds to the data set HairEyeColor in the datasets package,
Working With Categorical Variables
Categorical variables are usually represented as:
- character vectors
- factors.
Some advantages of factors:
- more control over ordering of levels
- levels are preserved when forming subsets
Most plotting and modeling functions will convert character vectors to factors with levels ordered alphabetically.
Some standard R functions for working with factors include
factorcreates a factor from another type of variablelevelsreturns the levels of a factorreorderchanges level order to match another variablerelevelmoves a particular level to the first position as a base linedroplevelsremoves levels not in the variable.
The tidyverse package forcats adds some more tools, including
fct_inordercreates a factor with levels ordered by first appearancefct_infreqorders levels by decreasing frequencyfct_revreverses the levelsfct_recodechanges factor levelsfct_relevelmoves one or more levelsfct_cmerges two or more factors
Bar Charts For Frequencies
Basics
The bar chart is often used to show the frequencies of a categorical variable.
By default, geom_bar uses stat = "count" and maps its result to the y aesthetic. This is suitable for raw data:
ggplot(raw) + geom_bar(aes(x = Hair))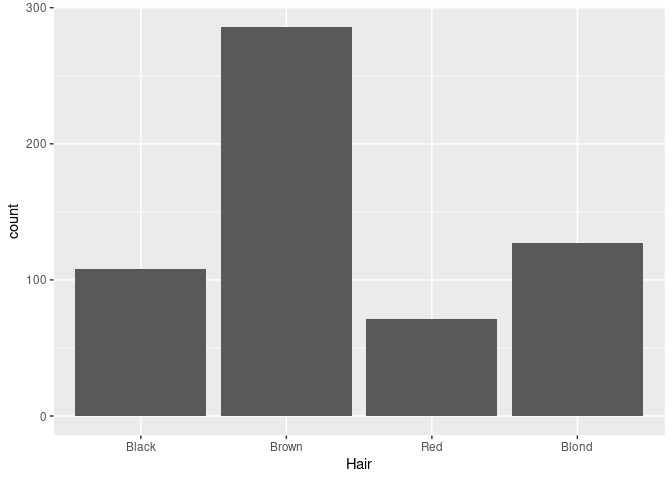
For a nominal variable it is often better to order the bars by decreasing frequency:
library(forcats)
ggplot(mutate(raw, Hair = fct_infreq(Hair))) + geom_bar(aes(x = Hair))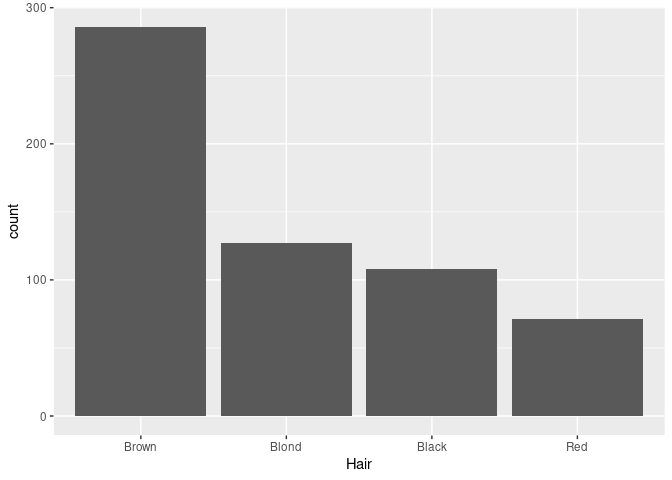
If the data have already been aggregated, then you need to specify stat = "identity" as well as the variable containing the counts as the y aesthetic:
ggplot(agg) + geom_bar(aes(x = Hair, y = n), stat = "identity")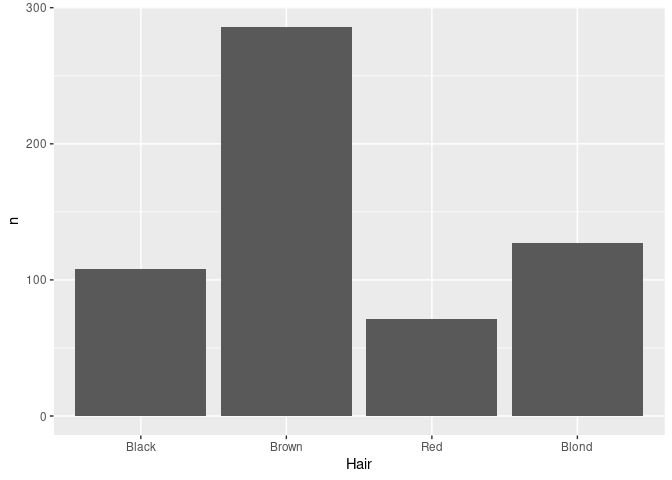
An alternative is to use geom_col.
For aggregated data reordering can be based on the computed counts using
agg_ord <- mutate(agg, Hair = reorder(Hair, -n, sum))-nis used to order largest to smallest;- the default summary used by
reorderismean;sumis better here.
ggplot(agg_ord) + geom_col(aes(x = Hair, y = n))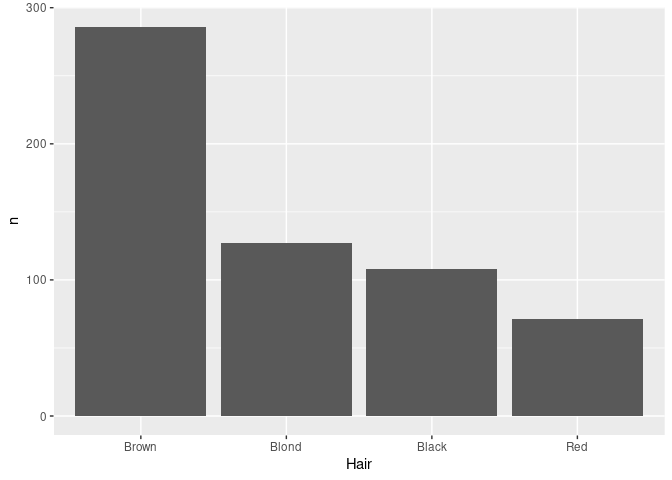
Adding a Grouping Variable
Mapping the Eye variable to fill in ggplot produces a stacked bar chart.
An alternative, specified with position = "dodge", is a side by side bar chart, or a clustered bar chart.
For the side by side chart in particular it may be useful to also reorder the Eye color levels.
ecols <- c(Brown = "brown2", Blue = "blue2",
Hazel = "darkgoldenrod3", Green = "green4")
agg_ord <- mutate(agg,
Hair = reorder(Hair, -n, sum),
Eye = reorder(Eye, -n, sum))
p1 <- ggplot(agg_ord) +
geom_col(aes(x = Hair, y = n, fill = Eye)) +
scale_fill_manual(values = ecols)
p2 <- ggplot(agg_ord) +
geom_col(aes(x = Hair, y = n, fill = Eye), position = "dodge") +
scale_fill_manual(values = ecols)
grid.arrange(p1, p2, nrow = 1)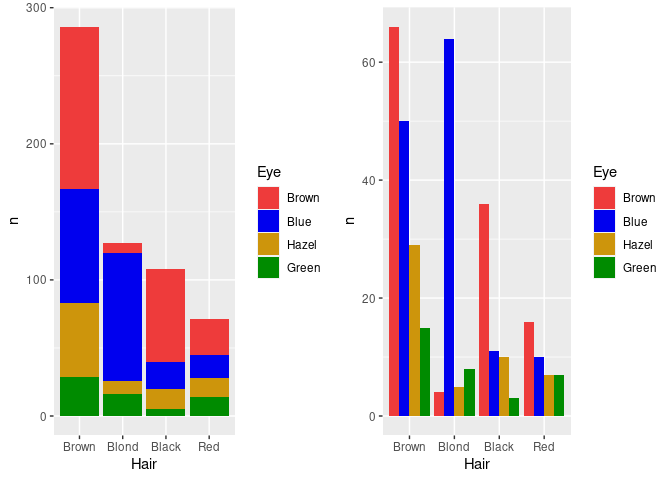
Faceting can be used to bring in additional variables:
p1 + facet_wrap(~ Sex)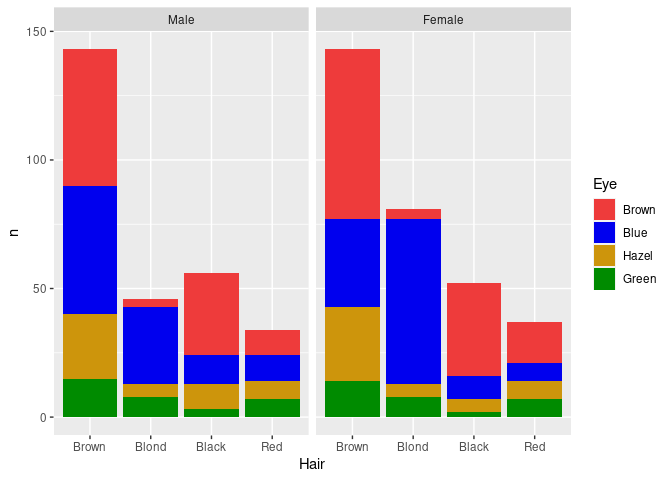
The counts shown here may not be the most relevant features for understanding the joint distributions of these variables.
Pie Charts and Doughnut Charts
Pie charts can be viewed as stacked bar charts in polar coordinates:
hcols <- c(Black = "black", Brown = "brown3",
Red = "brown1", Blond = "lightgoldenrod1")
p1 <- ggplot(agg_ord) +
geom_col(aes(x = 1, y = n, fill = Hair), position = "fill") +
coord_polar(theta = "y") +
scale_fill_manual(values = hcols)
p2 <- ggplot(agg_ord) +
geom_col(aes(x = Hair, y = n, fill = Hair)) +
scale_fill_manual(values = hcols)
grid.arrange(p1, p2, nrow = 1)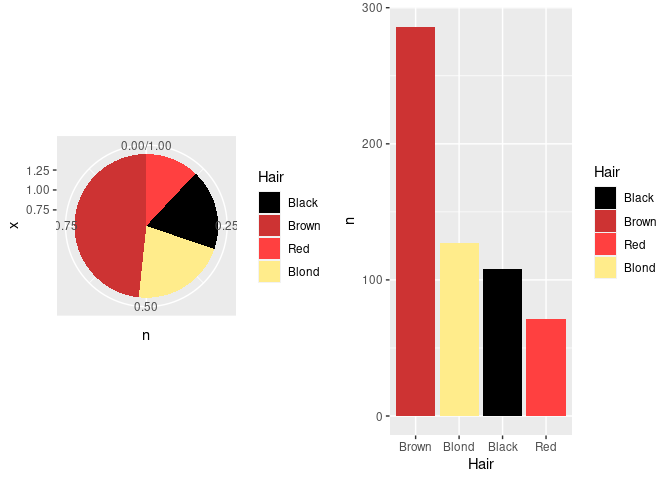
The axes and grid lines are not helpful for the pie chart and can be removed with some theme settings.
Using faceting we can also separately show the distributions for men and women:
p3 <- p1 + facet_wrap(~ Sex) +
theme_bw() +
theme(axis.title = element_blank(),
axis.text = element_blank(),
axis.ticks = element_blank(),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.border = element_blank())
p3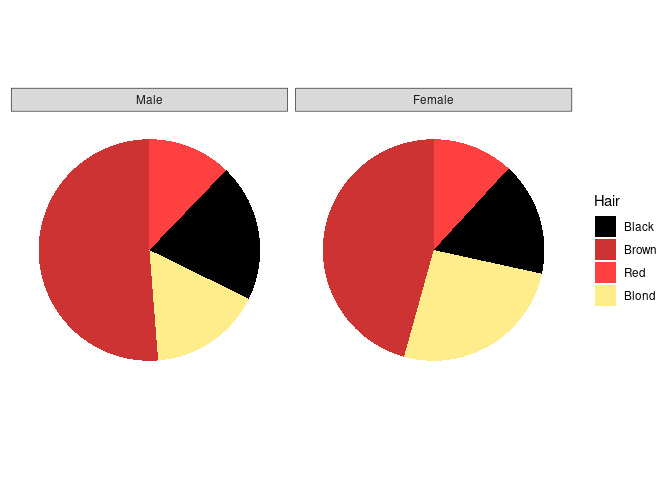
Doughnut charts are a variant that has recently become popular in the media:
p4 <- p3 + xlim(0, 1.5)
p4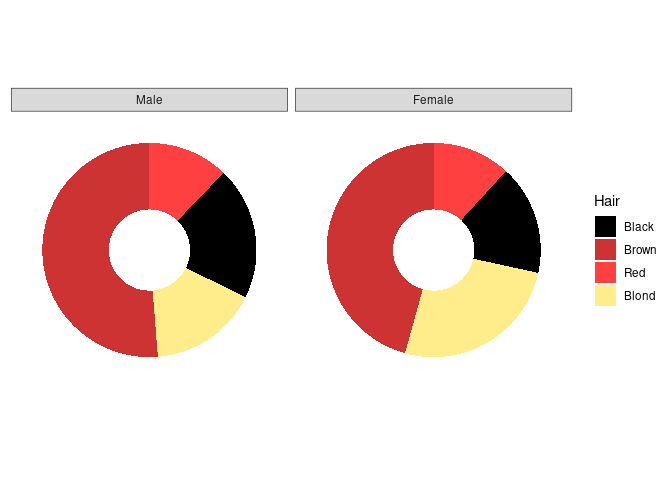
The center is often used for annotation:
p4 + geom_text(aes(x = 0, y = 0, label = Sex)) +
theme(strip.background = element_blank(),
strip.text = element_blank())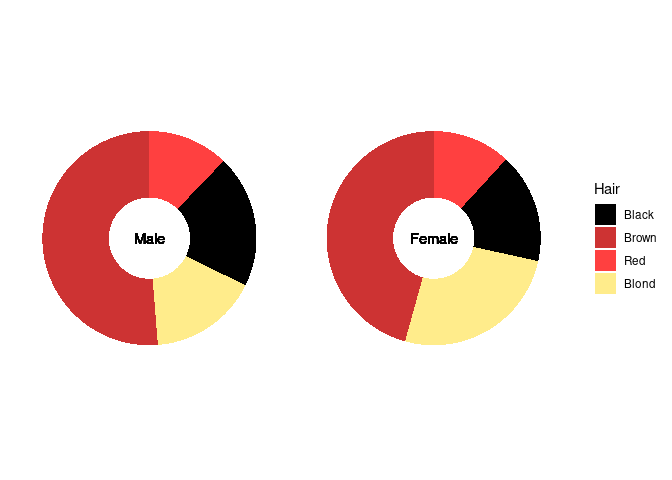
Some Notes
- Pie charts are effective for judging part/whole relationships.
- Pie charts are not very effective for comparing proportions.
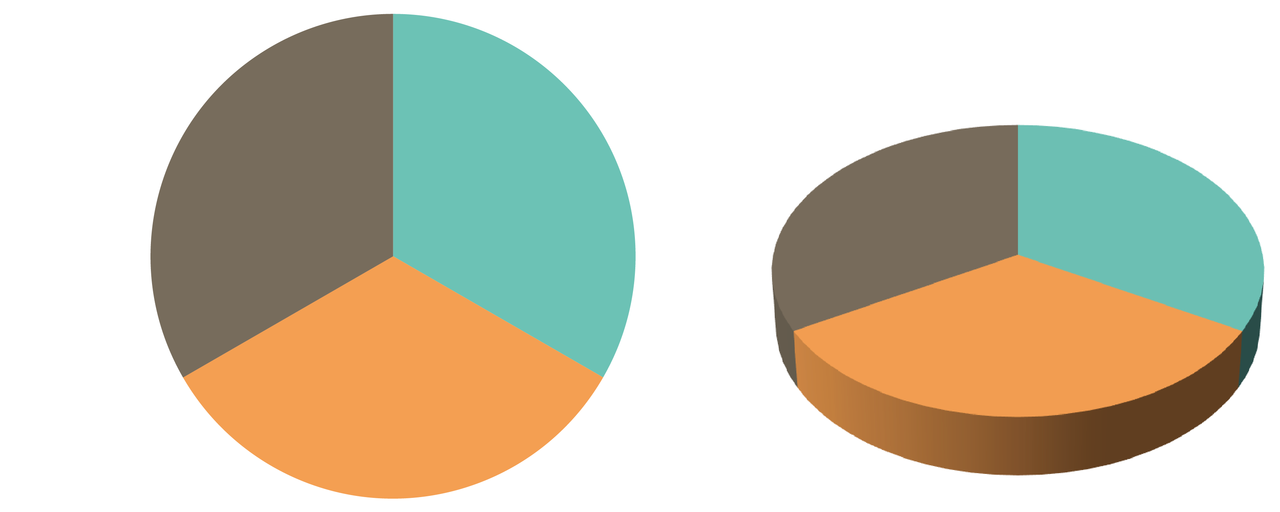
- 3D pie charts are popular and a very bad idea. An example (Fig. 6.61) from Andy Kirk’s book (2016), Data Visualization: A Handbook for Data Driven Design:
Stacked bar charts with equal heights are an alternative for representing part-whole relationhips:
ggplot(agg) +
geom_col(aes(x = Sex, y = n, fill = Hair), position = "fill") +
scale_fill_manual(values = hcols)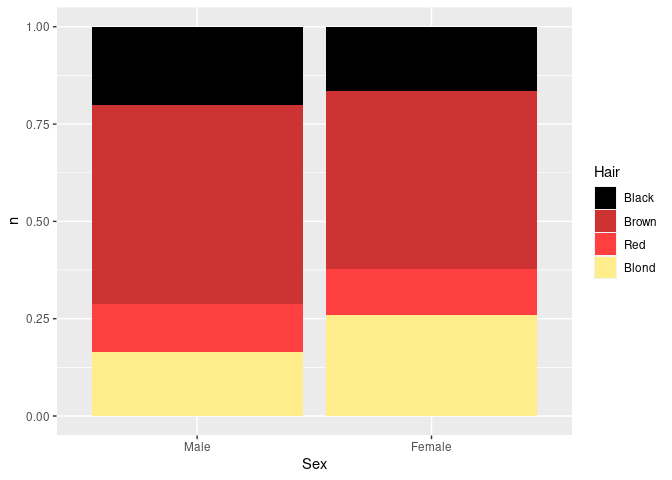
Another alternative is a waffle chart, sometimes also called a square pie chart.
The waffle package is one R implementation of this idea.
