More Examples
- More Global Average Surface Temperatures
- News cycle example
- Polar CO2 plot
- Measles heatmap
- Squirrels in central park
- Ridgeline map
- Minard’s Graph of Napoleon’s Russia Campaign
- Is Uber Replacing Taxis in Central Manhattan?
- Law of Large Numbers Simulation
- Vox Plot Reconstruction
- Weekly US Unemployment
More Global Average Surface Temperatures
if (! file.exists("GLB.Ts+dSST.csv"))
download.file("https://stat.uiowa.edu/~luke/data/GLB.Ts+dSST.csv",
"GLB.Ts+dSST.csv")
library(readr)
gast <- read_csv("GLB.Ts+dSST.csv", skip = 1)[1 : 13]
gast <- mutate_if(gast, is.character, as.numeric)
lgast <- pivot_longer(gast, -Year, names_to = "Month", values_to = "Temp") |>
mutate(Month = forcats::fct_inorder(Month))
if (class(lgast$Temp) == "character") {
lgast <- mutate(lgast, Temp = as.numeric(Temp))
filter(lgast, is.na(Temp))
}
p <- ggplot(lgast, aes(x = Month, y = Temp, group = Year)) +
ggtitle("Global Average Surface Temperatures") +
theme(plot.title = element_text(hjust = 0.5))We can use color to distingish the years.
p1 <- p + geom_line(aes(x = Month, y = Temp, group = Year, color = Year),
na.rm = TRUE)
p1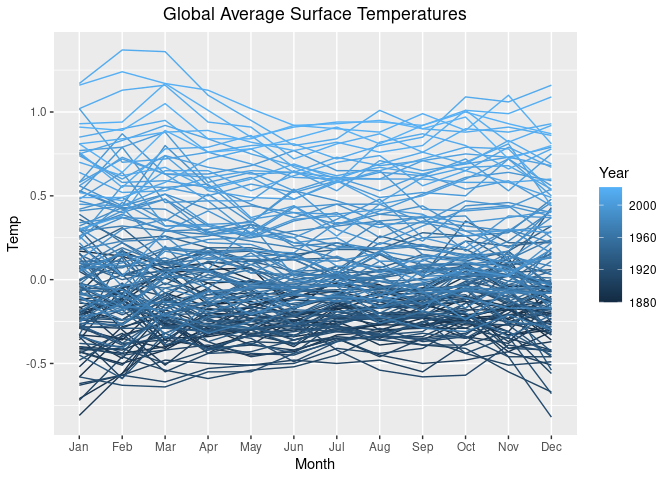
For a color gradient from black to red we can use
p1 + scale_color_gradient(low = "black", high = "red")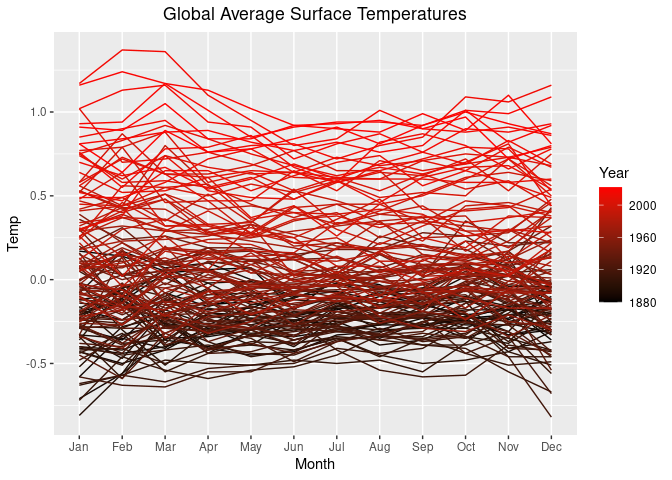
Another possibility is to use a continuous version of the "Reds" palette from https://colorbrewer2.org:
p2 <- p1 + scale_color_distiller(palette = "Reds", direction = 1)
p2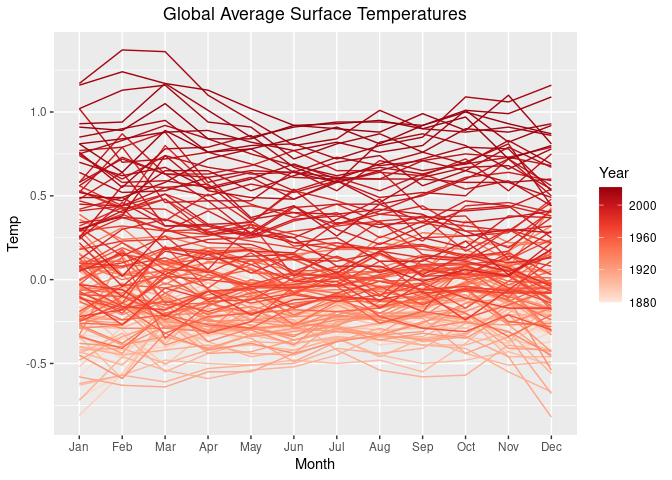
One way to highlight the past_year 2024:
lgast_last <- filter(lgast, Year == past_year)
p2 + geom_line(aes(x = Month, y = Temp, group = Year),
size = 1, color = "blue", data = lgast_last, na.rm = TRUE)
## Warning: Using `size` aesthetic for lines was deprecated in ggplot2 3.4.0.
## ℹ Please use `linewidth` instead.
## This warning is displayed once every 8 hours.
## Call `lifecycle::last_lifecycle_warnings()` to see where this warning was
## generated.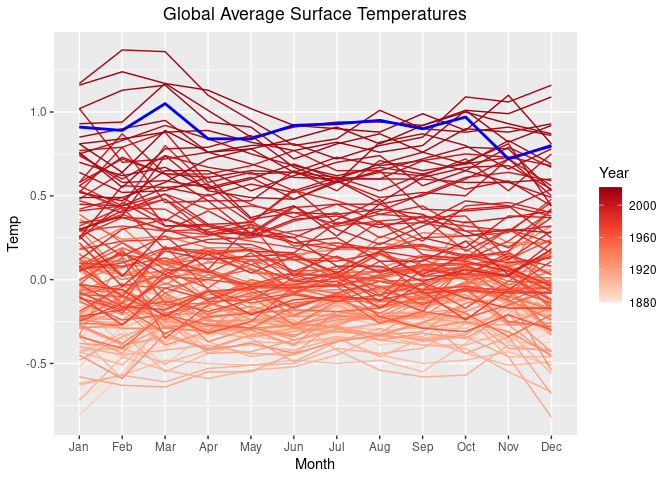
Show full data in gray and data since 2000 in black:
lgast2k <- filter(lgast, Year >= 2000)
ggplot(lgast, aes(x = Month, y = Temp, group = Year)) +
geom_line(color = "grey80") +
theme_minimal() +
geom_line(data = lgast2k)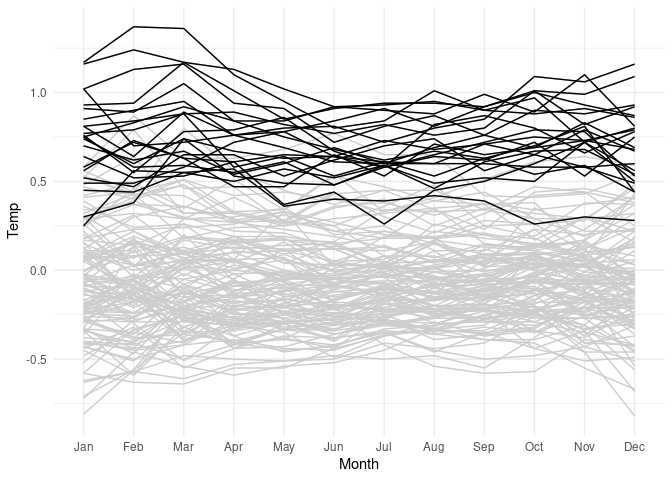
Show the years that set average temperature records:
mt <- group_by(lgast, Year) |>
summarize(mean_temp = mean(Temp)) |>
ungroup() |>
mutate(record = mean_temp == cummax(mean_temp))
lgast <- left_join(lgast, mt, "Year")
library(scales)
ggplot(lgast, aes(Month, Temp, group = Year)) +
geom_line(color = "grey") + theme_minimal() +
geom_line(aes(color = Year), data = filter(lgast, record)) +
scale_color_gradient2(midpoint = 1950,
low = muted("blue"),
high = muted("red"),
mid = muted("purple", l = 60))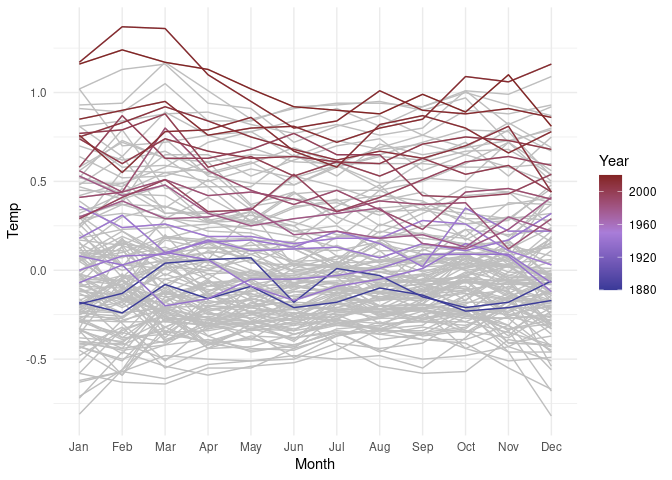
News cycle example
Slightly simplified example from Bob Rudis
library(ggalt)
library(hrbrthemes) # hrbrmstr/hrbrthemes
library(tidyverse)
jsonlite::fromJSON("https://rud.is/dl/2019-axios-news.json") |>
as_tibble() -> xdf
unnest(xdf, data) |>
group_by(name) |>
mutate(idx = seq_len(n())) |>
ungroup() |>
## make a factor for strip/panel ordering
mutate(name = fct_inorder(name)) -> xdf
sprintf("2019-%02s-1", 1 : 51) |>
as.Date(format = "%Y-%W-%w") |>
format("%b") |>
rle() -> mons
month_idx <- cumsum(mons$lengths) - 3
ggplot(xdf, aes(idx, data)) +
geom_area(aes(fill = avg, color = avg), alpha = 0.5) +
geom_hline(data = distinct(xdf, name, avg),
aes(yintercept = 0, colour = avg), size = 0.5) +
facet_wrap(~ name, ncol = 1, strip.position = "left", dir = "h") +
theme(strip.text.y = element_text(angle = 180, hjust = 1, vjust = 0),
legend.position = "none",
axis.text.y = element_blank(),
axis.ticks.y = element_blank(),
strip.background = element_blank(),
panel.spacing.y = unit(-0.5, "lines"),
panel.background = element_blank(),
panel.grid.major.y = element_blank(),
panel.grid.major.x = element_line(color = "grey80"),
panel.grid.minor.x = element_blank()) +
labs(x = NULL, y = NULL) +
scale_y_continuous(expand = c(0, 0), limits = c(0, 105)) +
scale_colour_viridis_c(option = "inferno", direction = -1,
begin = 0.1, end = 0.9) +
scale_fill_viridis_c(option = "inferno", direction = -1,
begin = 0.1, end = 0.9) +
scale_x_continuous(expand = c(0, 0.125), limits = c(1, 51),
breaks = month_idx, labels = month.abb)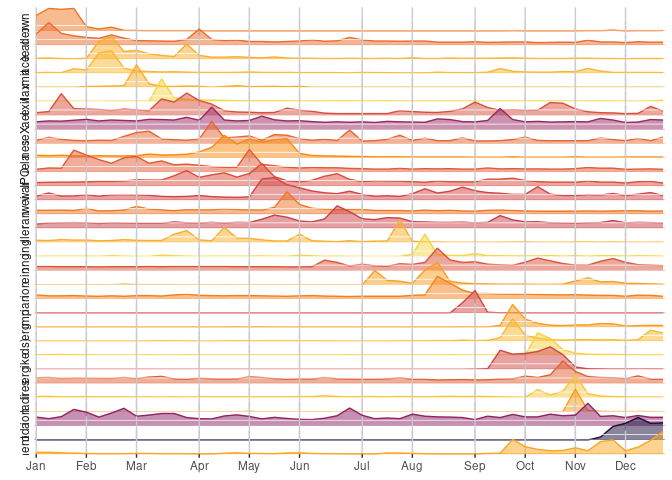
Polar CO2 plot
Mauna Loa CO2 data in polar coordinates:
https://r.iresmi.net/2020/01/01/mauna-loa-co2-polar-plot/
library(readr)
library(ggplot2)
library(dplyr)
co2ml <-
read_delim("ftp://aftp.cmdl.noaa.gov/products/trends/co2/co2_mm_mlo.txt",
delim = " ",
locale = locale(decimal_mark = "."),
na = c("-99.99", "-1"),
col_types = "iiddddi",
col_names = c("year", "month", "decimal", "co2",
"co2_interpol", "co2_trend", "days"),
comment = "#",
trim_ws = TRUE)
## Warning: One or more parsing issues, call `problems()` on your data frame for details,
## e.g.:
## dat <- vroom(...)
## problems(dat)
co2cir <- filter(co2ml, month == 1) |>
mutate(month = 12.999, year = year - 1)
co2cir <- rbind(co2cir, co2ml)
co2cir <- arrange(co2cir, year, month)
ggplot(co2cir, aes(month, co2_interpol, group = year, color = year)) +
geom_line() +
scale_color_viridis_c() +
scale_x_continuous(breaks = 1 : 12, labels = month.abb) +
coord_polar() +
labs(x = "",
y = CO[2] ~ "fraction in dry air (" * mu * "mol/mol)",
title = paste("Mauna Loa", min(co2ml$year), "-", max(co2ml$year)),
subtitle = "Monthly mean" ~ CO[2] ~ "concentration") +
theme_bw() +
theme(axis.title.y = element_text(hjust = .85),
panel.grid.major.y = element_blank(),
panel.grid.minor.x = element_blank(),
panel.border = element_blank())
## Warning: Removed 7 rows containing missing values or values outside the scale range
## (`geom_line()`).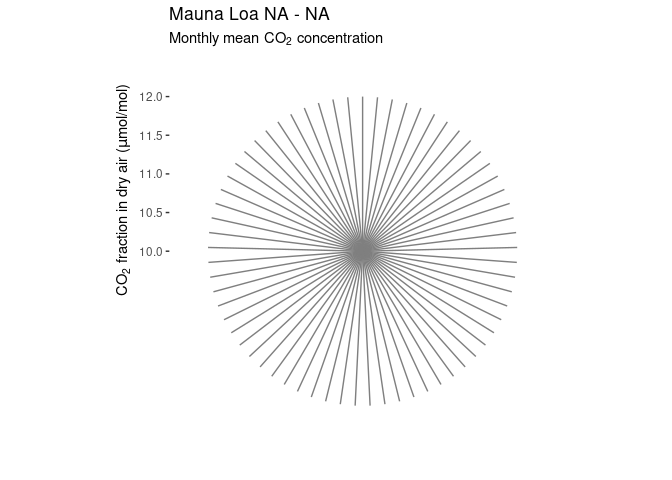
Measles heatmap
Rafa, p. 203
library(RColorBrewer)
library(dslabs)
library(ggplot2)
library(dplyr)
library(ggnewscale)
d <- mutate(us_contagious_diseases,
rate = 10000 * (count / population) * (52 / weeks_reporting)) |>
filter(! state %in% c("Alaska", "Hawaii") & disease == "Measles")
hm <- function(d)
ggplot(d) +
geom_tile(aes(x = year, y = state, fill = rate)) +
scale_x_continuous(expand = c(0, 0)) +
scale_fill_gradientn(colors = brewer.pal(9, "Reds"), trans = "sqrt") +
new_scale_fill() +
geom_tile(data = filter(d, is.na(rate)),
aes(x = year, y = state, fill = "NA")) +
scale_fill_manual(values = "grey50") +
geom_vline(xintercept = 1963, color = "blue") +
theme_minimal() +
theme(panel.grid = element_blank(),
legend.position = "bottom",
text = element_text(size = 8)) +
guides(fill = guide_legend(title = NULL, label.position = "bottom"))
hm(d)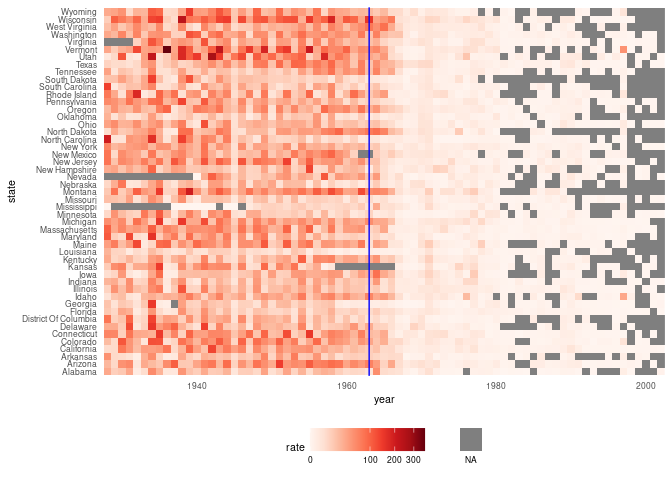
https://eliocamp.github.io/codigo-r/2018/09/multiple-color-and-fill-scales-with-ggplot2/
d_raw <- read_csv("US.14189004.csv",
col_types = cols(CityName = col_character(),
Admin2Name = col_character(),
PlaceOfAcquisition = col_character()))
d <- distinct(d_raw)
d <- filter(d, PartOfCumulativeCountSeries == 0)
d <- transmute(d,
state = str_to_title(Admin1Name),
start = PeriodStartDate,
count = CountValue,
city = CityName,
county = Admin2Name)
ds <- group_by(d, state, start) |>
summarize(count = sum(count)) |>
group_by(state, year = year(start)) |>
summarize(count = sum(count), weeks = n()) |>
ungroup()
ds <- left_join(ds, select(us_state_populations, -GISJOIN),
c("state", "year")) |>
fill(population) |>
complete(state, year)
lower48 <- setdiff(state.name, c("Hawaii", "Alaska"))
filter(ds, year >= 1930, state %in% lower48) |>
mutate(rate = 10000 * (count / population) * (52 / weeks)) |>
hmSquirrels in central park
https://flowingdata.com/2020/01/09/squirrel-census-count-in-central-park/ https://data.cityofnewyork.us/Environment/2018-Central-Park-Squirrel-Census-Squirrel-Data/vfnx-vebw
Ridgeline map
https://flowingdata.com/2020/01/06/draw-a-ridgeline-map-showing-elevation-for-anywhere-on-earth/
Base graphics:
v <- sweep(80 * volcano / max(volcano), 2, 1 : 61, FUN = `+`)
matplot(v, type = "l", col = "black", lty = 1)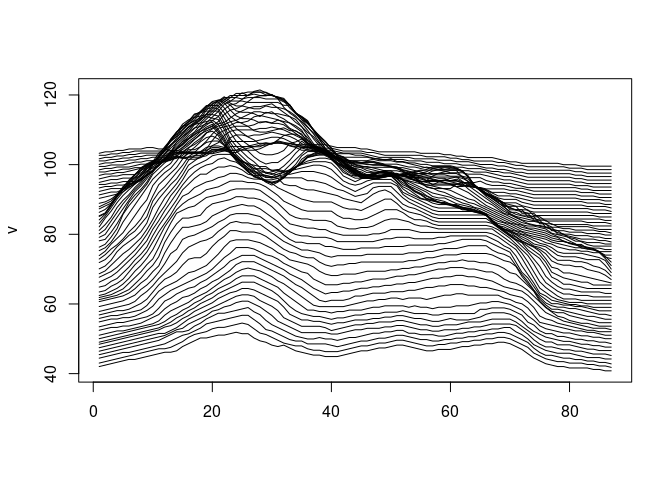
ggplot:
vd <- data.frame(value = as.numeric(volcano),
row = rep(seq_len(nrow(volcano)), ncol(volcano)),
col = rep(seq_len(ncol(volcano)), each = nrow(volcano)))
vd <- mutate(vd, y = 20 * (value / max(value)) + col)
p1 <- ggplot(vd, aes(x = row, y = y, group = rev(col))) +
geom_polygon(fill = "white", color = "black") +
theme_void()
p2 <- ggplot(vd, aes(x = row, ymax = y, ymin = min(y), group = rev(col))) +
geom_ribbon(fill = "white", color = "black") +
theme_void()
p0 <- ggplot(vd, aes(x = row, y = y, group = rev(col))) +
geom_line() +
theme_void()
p0
p1
p2
p0 + coord_fixed(ratio = ncol(volcano) / nrow(volcano))
p1 + coord_fixed(ratio = ncol(volcano) / nrow(volcano))
p2 + coord_fixed(ratio = ncol(volcano) / nrow(volcano))
Minard’s Graph of Napoleon’s Russia Campaign
Adapted from Rahlf’s book.
library(tidyverse)
library(HistData)
library(ggthemes)
all <- mutate(Minard.troops, hwidth = survivors / 1000000)
grp1 <- filter(all, group == 1)
grp2 <- filter(all, group == 2)
grp3 <- filter(all, group == 3)
p <- ggplot(NULL, aes(x = long,
ymin = lat - hwidth,
ymax = lat + hwidth,
fill = direction)) +
geom_ribbon(data = grp2[1 : 8, ]) +
geom_ribbon(data = grp2[8 : 10, ]) +
geom_ribbon(data = grp3) +
geom_ribbon(data = grp1) +
geom_text(aes(x = long, y = lat, label = city),
family = "Times New Roman", fontface = "italic", ##"Felipa",
data = Minard.cities, inherit.aes = FALSE) +
scale_fill_manual(values = c(A = "bisque2", R = "azure4")) +
coord_quickmap() +
theme_map() +
theme(panel.border = element_rect(fill = NA, colour = "grey20")) +
theme(legend.position = "bottom")
p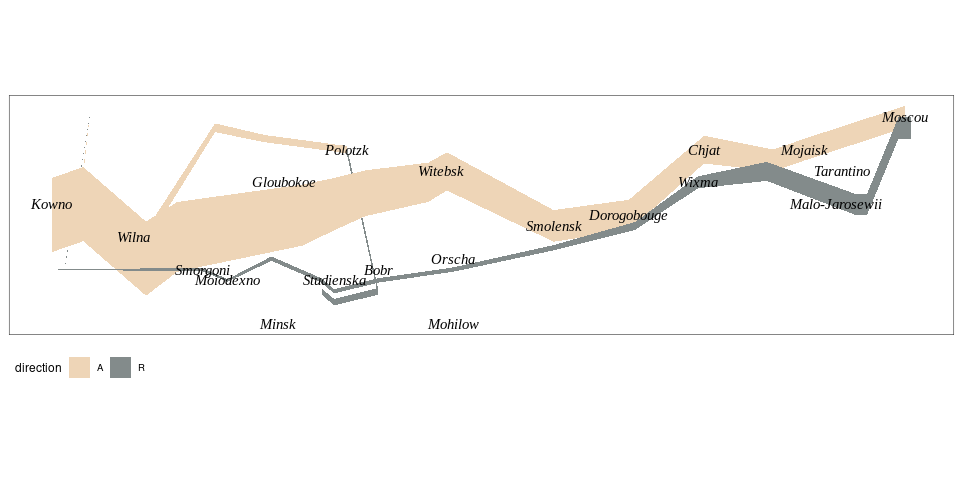
## May need this first
library(extrafont)
font_import()Adding the temperature graph:
temp_zero <- 53.5
temp_scale <- 30
temp_breaks <- c(-30, -20, -10, 0)
temps <- mutate(Minard.temp,
lat = temp_zero + temp / temp_scale,
temp_label = ifelse(is.na(date), temp, paste(temp, date)))
p + theme(legend.position = "top") +
geom_line(aes(x = long, y = lat), data = temps, inherit.aes = FALSE) +
geom_text(aes(x = long, y = lat, label = temp_label),
data = temps, nudge_y = -0.2, nudge_x = 0.1,
fontface = "italic", family = "Times New Roman", size = 3,
inherit.aes = FALSE) +
scale_y_continuous(position = "right",
breaks = temp_zero + temp_breaks / temp_scale,
labels = temp_breaks) +
theme(axis.text.y = element_text(),
axis.ticks.y = element_line(),
panel.grid.major.y = element_line(color = "grey80"))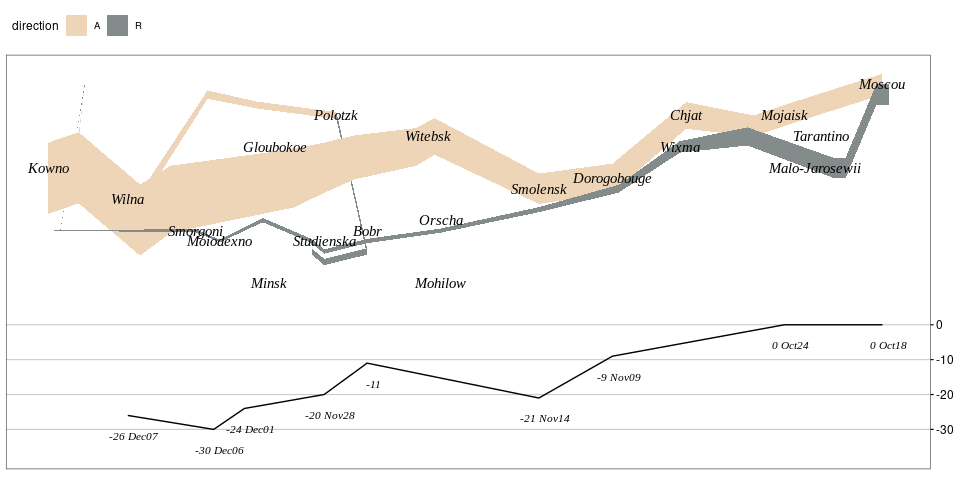
Is Uber Replacing Taxis in Central Manhattan?
Overall pickups among taxis, Uber, and similar services changed very little within central Manhattan from 2014 to 2015.
Taxi pickups decreased and Uber pickups increased. Was this a substitution of Uber for taxis?
An article on the FiveThirtyEight examines this question with a graph:
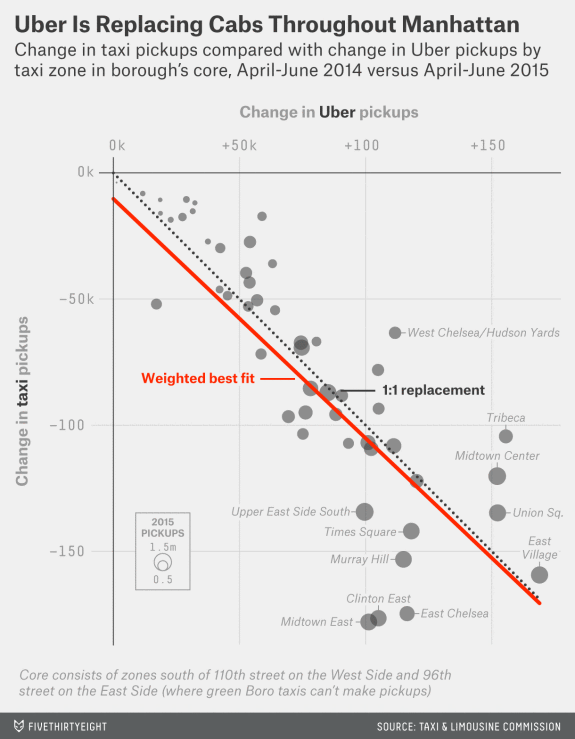
- Items: Taxi zones in Mahattan’s core.
- Attributes/variables:
- change in taxi pickups
- change in Uber pickups
- total number of pickups in the 2015 period
- Marks: points.
- Channels:
- horizontal position, mapped to Uber change
- vertical position, mapped to taxi change
- point size, mapped to total number of pickups
- Supporting features:
- Negative 45 degree line
- Regression line
- Annotations
An approximate reconstruction using an approximation to the data in the graph:
if (! file.exists("nyuber.dat"))
download.file("https://stat.uiowa.edu/~luke/data/nyuber.dat",
"nyuber.dat")
tu <- read.table("nyuber.dat", head = TRUE)
ggplot(tu, aes(x = uber, y = taxi)) +
geom_point(aes(size = rides)) +
geom_abline(slope = -1, linetype = 2) +
geom_smooth(method = "lm", se = FALSE, color = "red") +
scale_size_area()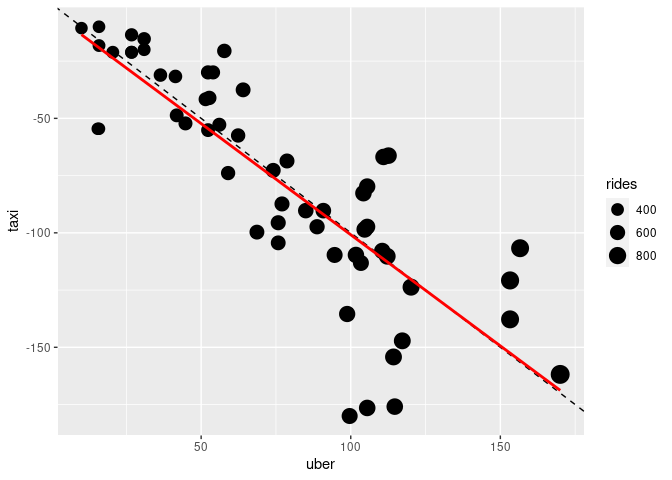
The primary question is to what degree at the zone level decreases in taxi pickups match increases in Uber pickups.
The strongest channels, 2D position, have been used for the two change variables.
Answering the question requires assessing the position of a point relative to the 1:1 replacement line. This is much harder than assessing position relative to an axis.
An alternate approach is to plot the number of taxi pickups lost per Uber ride gained against uber gains or taxi losses:
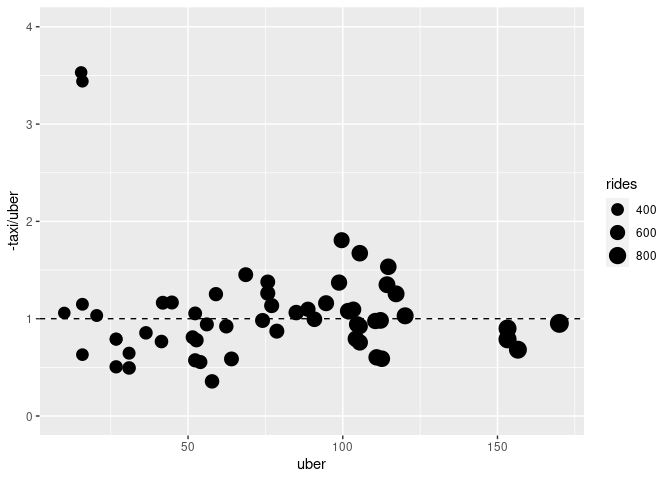
This allows the taxi/tradeoff to be assessed by comparison to a common scale.
Several other variations are possible.
Law of Large Numbers Simulation
Automated choices of axis scaling can affect the aspect ratio of the content of a plot.
A simple simulation of coins flips to illustrate the law of large numbers:
n <- 100000
prob <- 0.5
flips <- data.frame(trial = 1 : n, flip = rbinom(n, 1, prob))
library(dplyr)
flips <- mutate(flips, phat = cumsum(flip) / trial)
head(flips, 10)
## trial flip phat
## 1 1 1 1.0000000
## 2 2 1 1.0000000
## 3 3 0 0.6666667
## 4 4 1 0.7500000
## 5 5 1 0.8000000
## 6 6 0 0.6666667
## 7 7 0 0.5714286
## 8 8 0 0.5000000
## 9 9 1 0.5555556
## 10 10 1 0.6000000p <- ggplot(flips, aes(x = trial, y = phat)) +
geom_line() +
geom_abline(intercept = prob, slope = 0, lty = 2)
p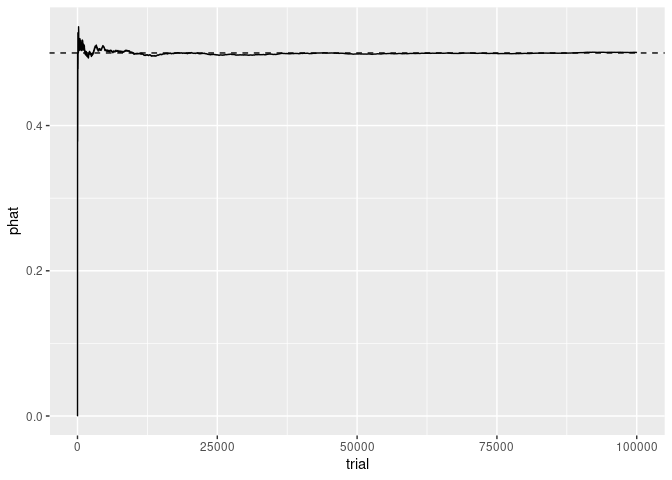
Focusing the y axis around 1/2 shows the early fluctuations near 1/2 more clearly:
p + coord_cartesian(ylim = c(0.45, 0.55))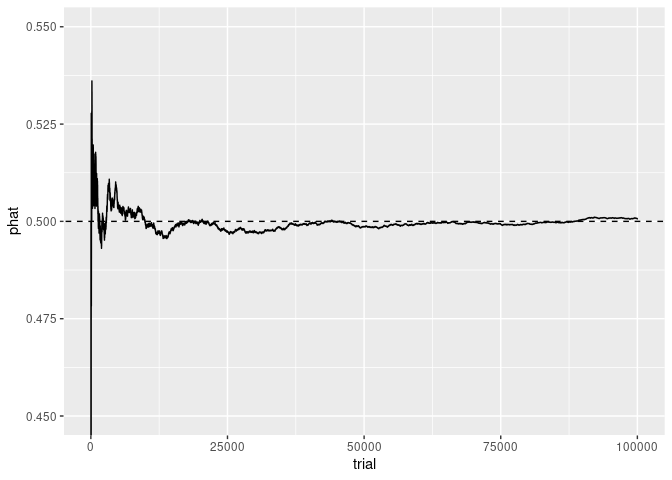
p + ylim(c(0.45, 0.55))
## Warning: Removed 7 rows containing missing values or values outside the scale range
## (`geom_line()`).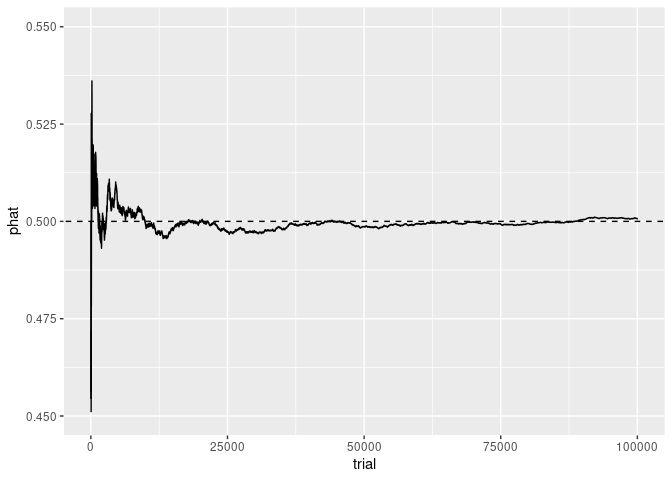
Focusing on the x axis for the last half of the trials shows that, as expected, the sample proportion is very close to 1/2.
The y axis scale still reflects the range of the complete data.
p + coord_cartesian(xlim = c(50000, 100000))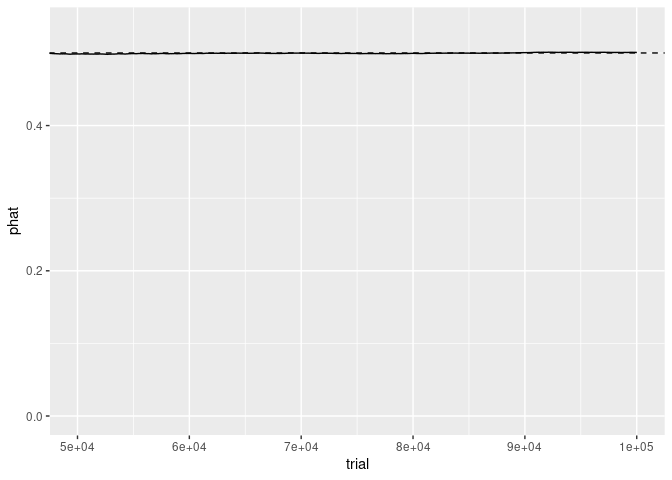
Using a subset of the data for trials >= 50000 produces a very different picture that emphasizes the details of the fluctuations:
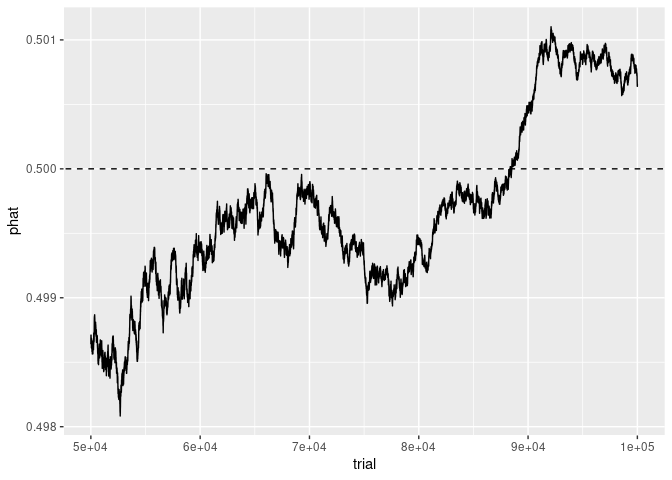
Vox Plot Reconstruction
Reconstruction of the revised Vox chart in the article on the ice bucket challenge.
library(ggplot2)
d <- read.csv("https://stat.uiowa.edu/~luke/data/dfunds.csv")
## dd <- data.frame(var = rep(c("Money Raised", "Deaths"), each = nrow(d)),
## val = c(d$Funding, d$Deaths / 1000),
## rank = c(rank(d$Funding), rank(d$Deaths)),
## Disease = rep(d$Disease, 2))
dd <- mutate(d, Deaths = Deaths / 1000) |>
rename("Money Raised" = Funding) |>
pivot_longer(-Disease, names_to = "var", values_to = "val") |>
group_by(var) |>
mutate(rank = rank(val)) |>
ungroup()
ggplot(dd, aes(x = var, y = rank)) +
geom_line(aes(group = Disease)) +
geom_point(aes(color = Disease, size = val)) +
geom_text(aes(label = round(val),
hjust = ifelse(var == "Deaths", "left", "right"),
x = ifelse(var == "Deaths",
2 + 0.01 + sqrt(val) / 120,
1 - 0.01 - sqrt(val) / 120))) +
scale_size_area(max_size = 25) +
scale_x_discrete(position = "top",
limits = c("Money Raised", "Deaths")) +
scale_y_continuous(expand = c(0, 0), limits = c(0, 9)) +
theme_minimal() +
theme(axis.title = element_blank(),
axis.text.y = element_blank(),
axis.ticks.y = element_blank(),
text = element_text(size = 15, face = "bold"),
legend.position = "top") +
guides(color = guide_legend(override.aes = list(size = 5),
nrow = 4,
title = NULL),
size = "none")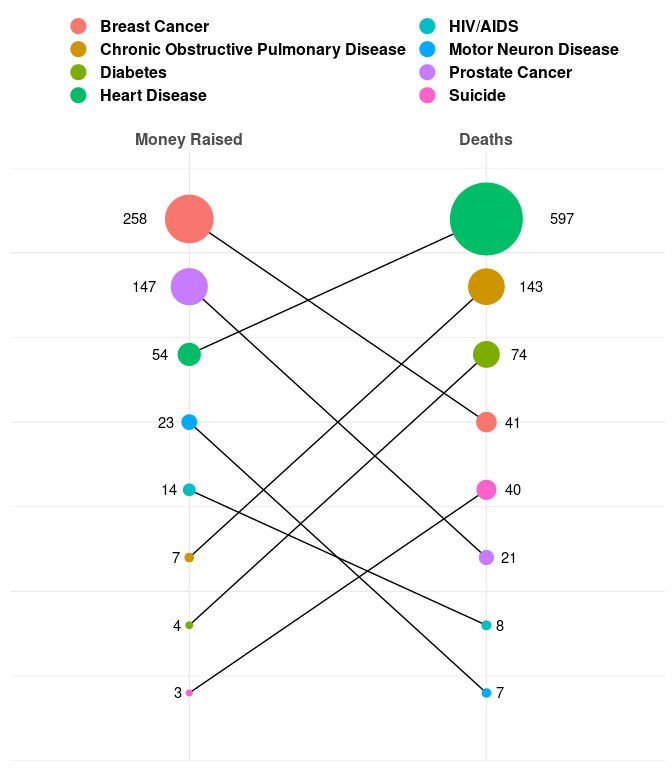
Variant:
d1 <- mutate(d,
var = "Deaths",
val = Deaths / 1000,
rank = rank(Deaths))
d2 <- mutate(d,
var = "Money Raised",
val = Funding,
rank = rank(Funding))
ggplot(NULL, aes(x = var, y = rank, size = val, color = Disease)) +
geom_point(data = d2) +
geom_point(data = d1) +
scale_size_area(max_size = 25, guide = "none") +
scale_x_discrete(name = NULL,
position = "top",
limits = c("Money Raised", "Deaths")) +
scale_y_continuous(name = NULL,
expand = c(0, 0),
limits = c(0, 9))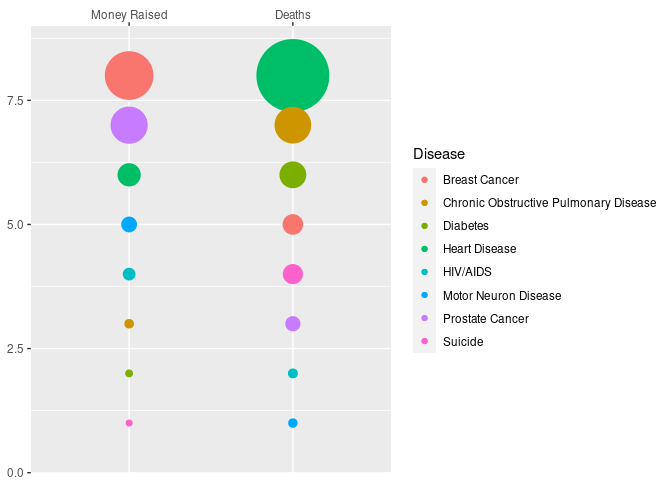
Can also ggforce::geom_circle. More painful but a nicer looking result.
library(ggforce)
gap <- 0.1
centr <- function(r) {
n <- length(r)
sr <- sort(r)
b <- cumsum(c(0, gap + 2 * sr[-n]))
(b + sr)[rank(r)]
}
ddr <- mutate(dd,
r = sqrt(val / max(val)),
x0 = ifelse(var == "Deaths", 3, 1)) |>
group_by(var) |>
mutate(y0 = centr(r)) |>
ungroup()
ddrr <- group_by(ddr, rank) |> mutate(y0 = max(y0))
ggplot(ddrr) +
geom_circle(aes(x0 = x0, y0 = y0 + gap, r = r, fill = Disease)) +
coord_fixed() +
scale_x_continuous(position = "top",
breaks = c(1, 3),
labels = c("Money Raised", "Deaths"),
expand = c(0.8, 0.8)) +
scale_y_continuous(expand = c(0, 0)) +
theme_minimal() +
theme(panel.grid = element_blank(),
legend.position = "top",
axis.title = element_blank(),
axis.text.y = element_blank(),
axis.ticks.y = element_blank(),
text = element_text(size = 15, face = "bold"),
plot.background = element_rect(fill = "beige")) +
guides(fill = guide_legend(nrow = 4,
title = NULL,
override.aes = list(color = NA)))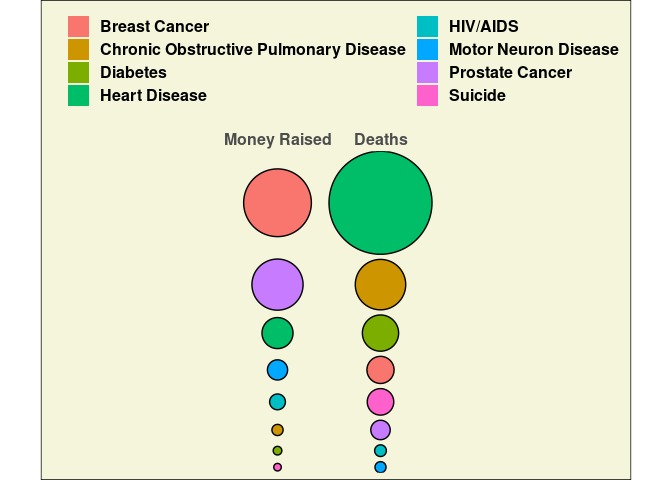
Weekly US Unemployment
I have not yet been able to find county level weekly unemployment data.
State level data is available from the US Department of Labor via a web interface or a page for downloading CSV files.
The most appropriate data is ETA 539. A skeletal variable description file is available.
Based on that description file and the data from the web interface the useful variables seem to be:
| Variable | Meaning |
|---|---|
st |
State |
rptdate F |
iled Week Ended |
c2 |
Reflecting Week Ended |
c3 |
Initial Claims |
c8 |
Continued Claims |
c18 |
Covered Employment |
c19 |
Insured Unemployment Rate |
Read and clean the data.
## **** move this to top of file
options(conflicts.policy = list(depends.ok = TRUE, warn = FALSE))
suppressMessages(library(tidyverse))
library(lubridate)
unemp_file <- "ar539.csv"
unemp_url <- "https://oui.doleta.gov/unemploy/csv/ar539.csv"
if (! file.exists(unemp_file) ||
now() > file.mtime(unemp_file) + weeks(1))
download.file(unemp_url, unemp_file)
ud <- read.csv(unemp_file)
ud <- transmute(ud,
state = st,
weekEnded = mdy(rptdate),
ReflectingWeekEnded = mdy(c2),
InitialClaims = c3,
ContinuedClaims = c8,
CoveredEmployment = c18,
InsuredUnemploymentRate = c19)Some time series:
filter(ud, state %in% c("IA", "IL", "WI", "MN"),
ReflectingWeekEnded > "2019-11-01") |>
ggplot(aes(x = ReflectingWeekEnded,
y = InsuredUnemploymentRate,
color = state)) +
geom_line()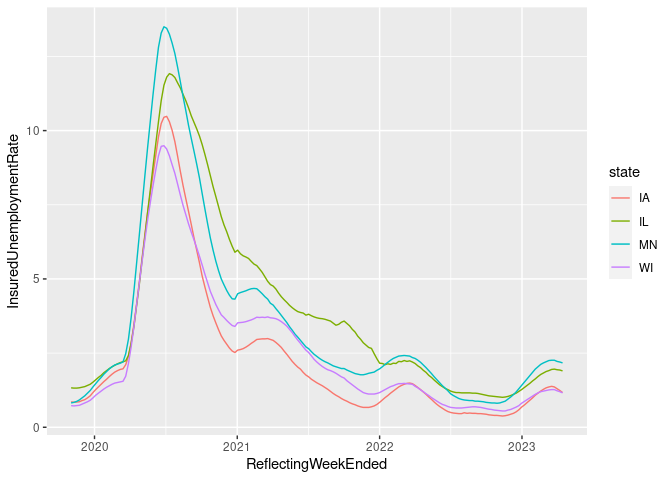
Choropleth maps for the last 6 weeks:
states_sf <- sf::st_as_sf(maps::map("state", plot = FALSE, fill = TRUE),
csr = 4326)
ud1 <- rename(ud, ID = state) |>
mutate(ID = tolower(tolower(state.name[match(ID, state.abb)])))
udf_sf <- mutate(states_sf, ID = as.character(ID)) |> left_join(ud1, "ID")
filter(udf_sf, weekEnded >= max(weekEnded, na.rm = TRUE) - weeks(5)) |>
rename(Rate = InsuredUnemploymentRate) |>
ggplot(aes(fill = Rate)) +
geom_sf() +
coord_sf() +
facet_wrap(~weekEnded, ncol = 2) +
scale_fill_distiller(palette = "Reds", direction = 1)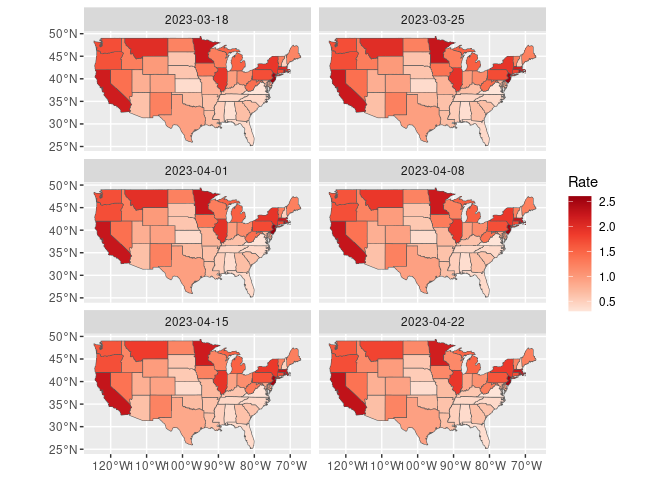
For the most recent period break out north-east states:
library(sf)
udf2_sf <- filter(udf_sf, weekEnded == max(weekEnded, na.rm = TRUE)) |>
rename(Rate = InsuredUnemploymentRate)
NE <- c("maine", "vermont", "new hampshire", "massachusetts",
"rhode island", "connecticut")
NE_sf <- filter(udf2_sf, ID %in% NE)
NE2_sf <- st_set_crs(st_set_geometry(NE_sf,
(st_geometry(NE_sf) - c(-71, 42.3)) * 3 + c(-50, 30)),
4326)
ggplot(NULL, aes(fill = Rate)) +
geom_sf(data = NE2_sf) +
geom_sf(data = filter(udf2_sf, ! ID %in% NE)) +
geom_sf(data = mutate(NE_sf, Rate = NULL), fill = NA) +
coord_sf() +
scale_fill_distiller(palette = "Reds", direction = 1)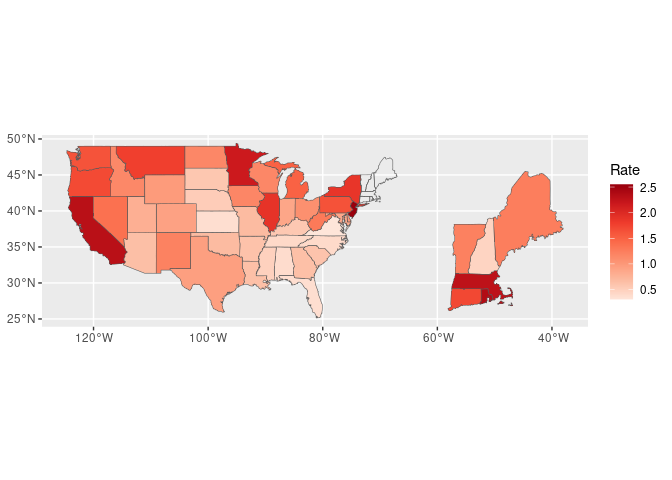
Unemployment time series spark line plot motivated by van example for France.
sf::sf_use_s2(FALSE)
states_sf <- cbind(states_sf, st_coordinates(st_centroid(states_sf)))
## Warning: st_centroid assumes attributes are constant over geometries
## Warning in st_centroid.sfc(st_geometry(x), of_largest_polygon =
## of_largest_polygon): st_centroid does not give correct centroids for
## longitude/latitude data
sf::sf_use_s2(TRUE)
start <- "2020-01-01"
udf3_sf <-
mutate(states_sf, ID = as.character(ID)) |>
left_join(filter(ud1, weekEnded >= start), "ID") |>
mutate(weeks = as.integer(difftime(weekEnded, ymd(start), units = "weeks")))
ggplot(states_sf) +
geom_sf(color = "grey") +
geom_line(aes(X + 0.02 * weeks,
Y + 0.1 * InsuredUnemploymentRate,
group = ID),
data = udf3_sf) +
ggtitle(paste("Unemployment Rates Since", start)) +
ggthemes::theme_map()
## Warning: Removed 1 row containing missing values or values outside the scale range
## (`geom_line()`).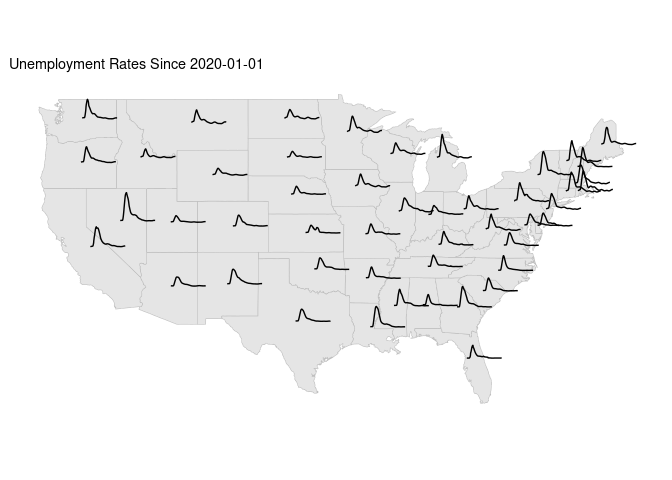
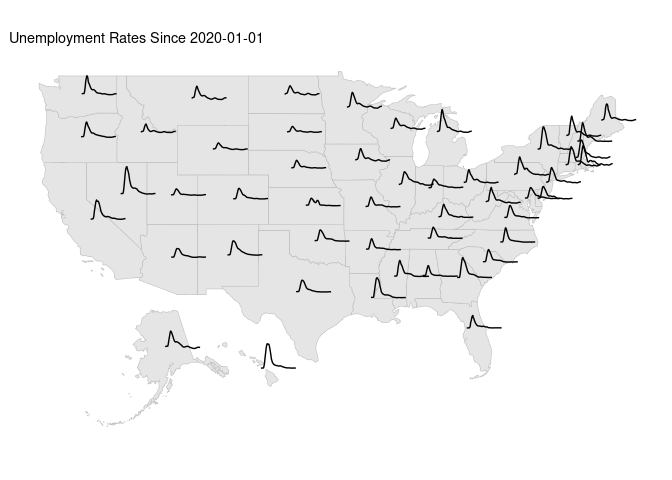
A variant including Alaska and Hawaii:
library(usmap)
usa_ha = us_map()
usa_ha <- cbind(usa_ha, st_coordinates(st_centroid(usa_ha)))
## Warning: st_centroid assumes attributes are constant over geometries
udf4_sf <-
mutate(usa_ha, ID = tolower(as.character(full))) |>
left_join(filter(ud1, weekEnded >= start), "ID") |>
mutate(weeks = as.integer(difftime(weekEnded, ymd(start), units = "weeks")))
ggplot(usa_ha) +
geom_sf(color = "grey") +
geom_line(aes(X + 2000 * weeks,
Y + 10000 * InsuredUnemploymentRate,
group = ID),
data = udf4_sf) +
ggtitle(paste("Unemployment Rates Since", start)) +
ggthemes::theme_map()
## Warning: Removed 1 row containing missing values or values outside the scale range
## (`geom_line()`).